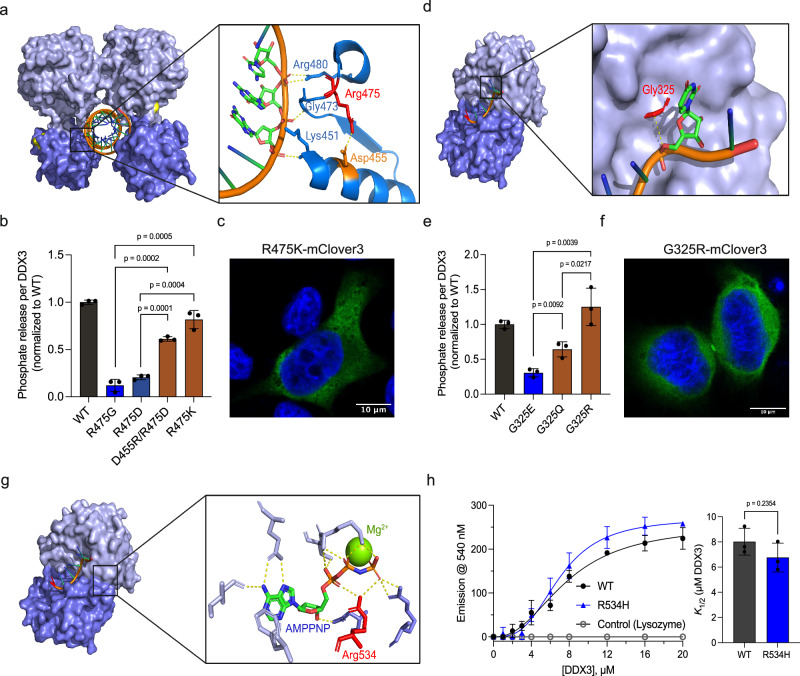
Fig. 3. Mechanistic studies reveal how DDX3X mutants interfere with catalysis.
a Pre-unwound structure of DDX3X zoomed on Arg475 (red) and the surrounding RNA-binding residues. Asp455 is noted in orange, and the dashed yellow line indicates a putative salt bridge between these residues. Their respective yellow dashed lines indicate interactions between Lys451, Gly473, and Arg480 and the RNA backbone. b ATPase activity of mCherry-tagged WT DDX3X, R475G, R475D, D455R/R475D, and R475K (normalized to WT). Significance was calculated using a two-tailed t-test. Values represent mean ± s.d., n = 3 independent reactions. c A representative image of R475K-mClover3 in HeLa cells, scale bar = 10 μm. d Post-unwound structure of Vasa zoomed in on Gly325 (red, numbered according to DDX3X homology). The dashed line indicates the 3.6 Å distance between the alpha carbon of Gly325 and the RNA backbone. e ATPase activity of mCherry-tagged WT DDX3X, G325E, G325Q, and G325R normalized to WT. Significance was calculated using a two-tailed t-test. Values represent mean ± s.d., n = 3 independent reactions. f A representative image of G325R-mClover3 in HeLa cells, scale bar = 10 μm. g Post-unwound structure of DDX3X zoomed in on the ATP-binding pocket, showing residues involved in ATP binding and Mg2+ cation (yellow sphere). Arg534 (numbered with DDX3X homology) is indicated in red. Yellow dashed lines indicated interactions between residues and ATP. h TNP-ATP binding curve for MBP-tagged WT DDX3X, R534H, and negative control (lysozyme), fit as described in methods. Right, K1/2 values for WT and R534H. Significance was calculated using a two-tailed t-test. Values represent mean ± s.e.m., n = 3 independent measurements. Source data are provided as a Source Data file.