Abstract
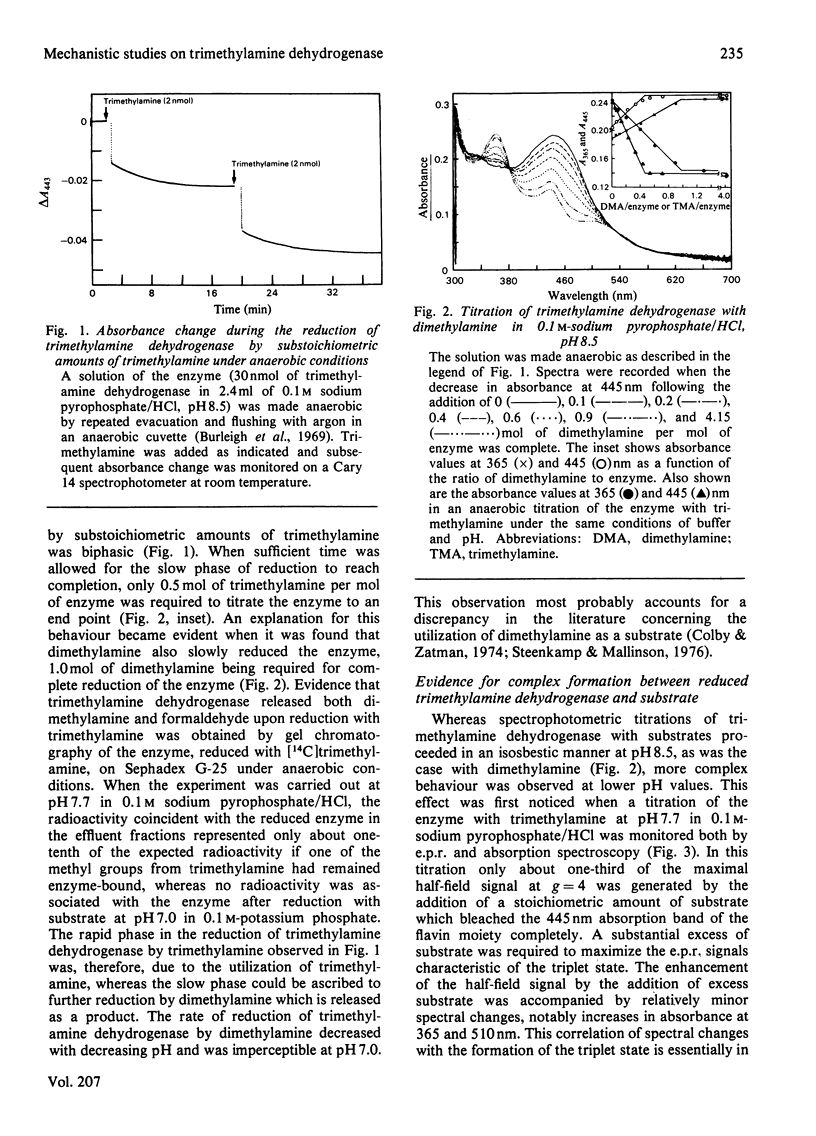
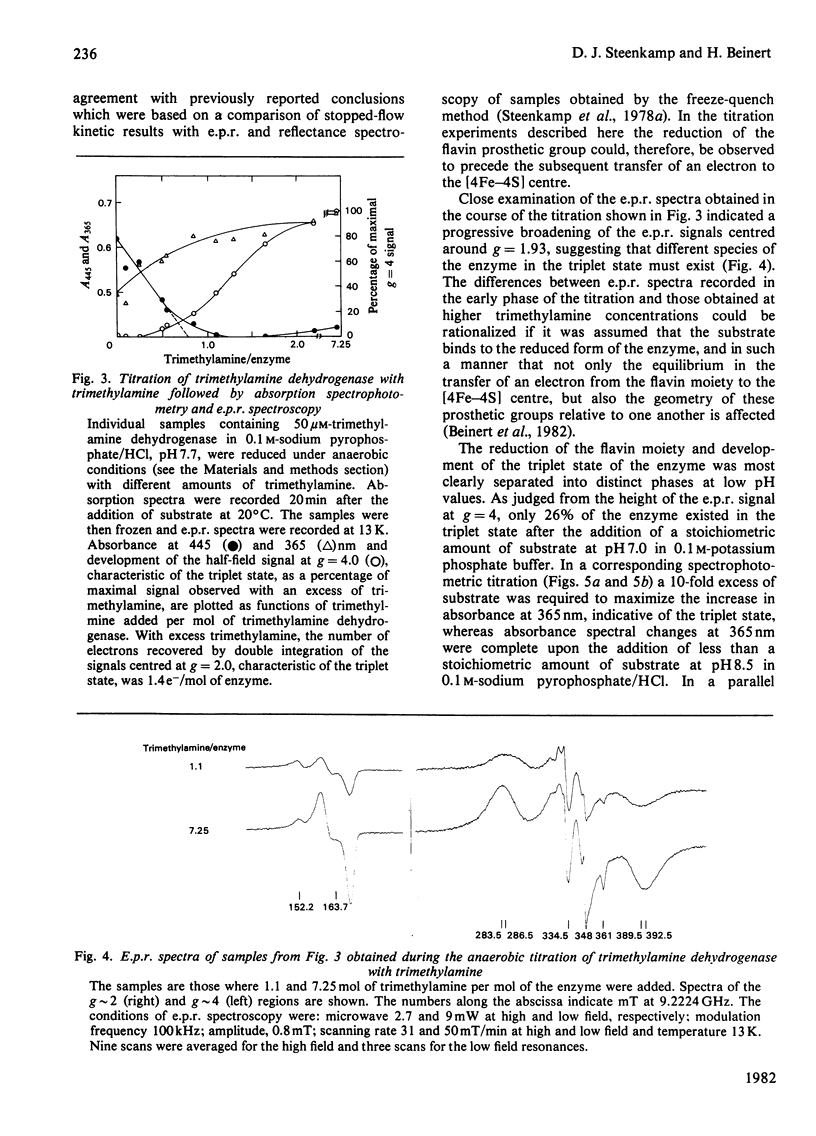
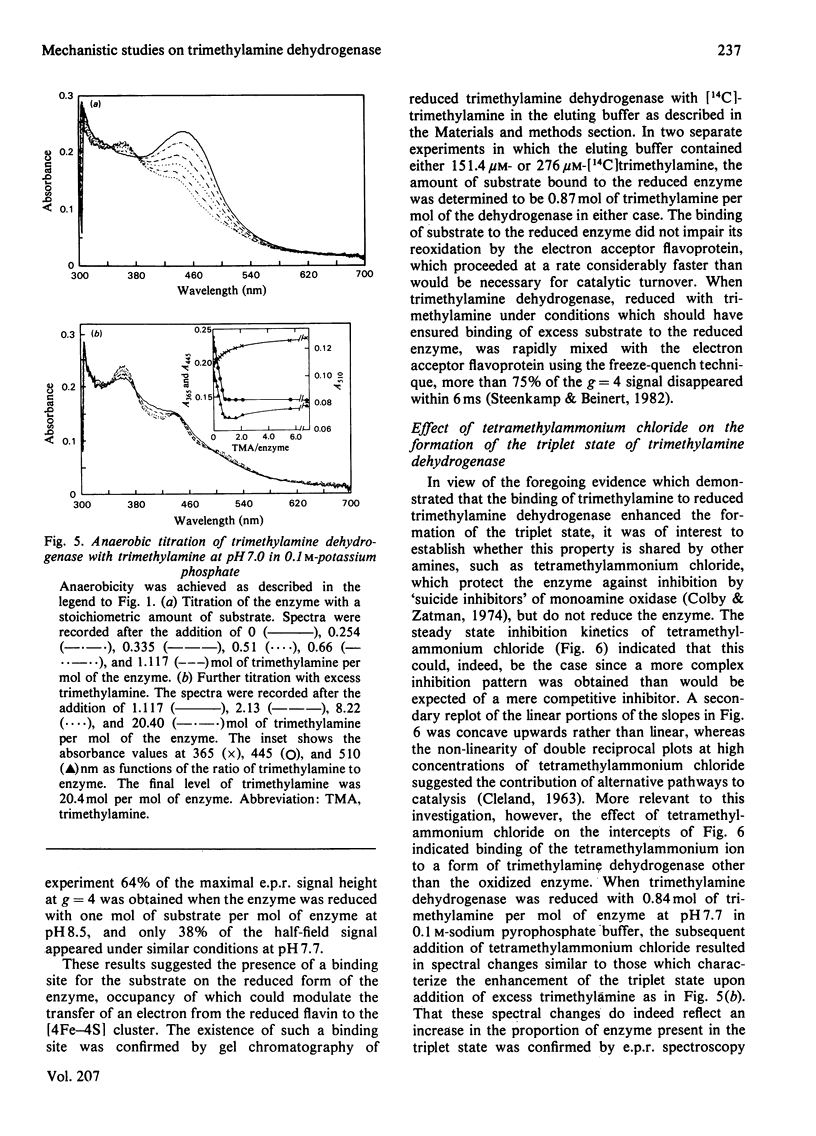
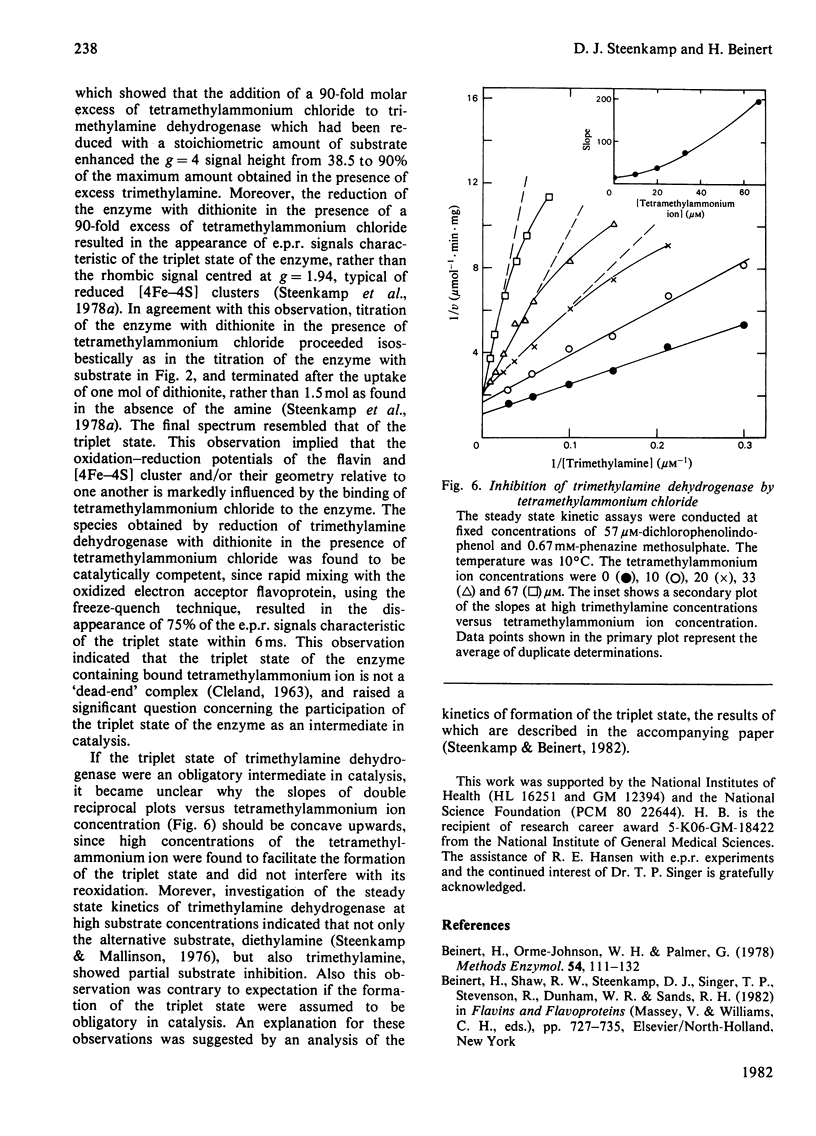
The trimethylamine dehydrogenase of bacterium W3A1 is reduced with the formation of a triplet state in which two electrons, derived from the substrate, are distributed between the [4Fe-4S] cluster and 6-S-cysteinyl-FMN semiquinone. In titration experiments at pH 8.5 about 1.0 mol of dimethylamine or 0.5 mol of trimethylamine per mol of the enzyme is required to titrate the enzyme to an endpoint. At pH values less than 8.0, however, an excess of trimethylamine is required to obtain maximal yield of the g = 4 e.p.r. signal, characteristic of the triplet state, or maximal absorbance at 365 nm which indicates formation of the flavin semiquinone. The binding of 0.86 mol of trimethylamine per mol of the enzyme could be detected by a gel chromatographic method. When the enzyme is titrated with dithionite in the presence of tetramethylammonium chloride, an endpoint is reached after the uptake of two electrons which give rise to the triplet state, whereas three electrons are consumed in the absence of tetramethylammonium chloride to reduce the enzyme completely. The enzyme is inhibited noncompetitively by tetramethylammonium chloride and the slopes of double reciprocal plots are a concave upwards function of inhibitor concentration. The data indicate the presence of a binding site for the substrate and other amines on the reduced enzyme which enhances the proportion of enzyme in the triplet state.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beinert H., Orme-Johnson W. H., Palmer G. Special techniques for the preparation of samples for low-temperature EPR spectroscopy. Methods Enzymol. 1978;54:111–132. doi: 10.1016/s0076-6879(78)54013-5. [DOI] [PubMed] [Google Scholar]
- Burleigh B. D., Jr, Foust G. P., Williams C. H., Jr A method for titrating oxygen-sensitive organic redox systems with reducing agents in solution. Anal Biochem. 1969 Mar;27(3):536–544. doi: 10.1016/0003-2697(69)90067-0. [DOI] [PubMed] [Google Scholar]
- CLELAND W. W. The kinetics of enzyme-catalyzed reactions with two or more substrates or products. II. Inhibition: nomenclature and theory. Biochim Biophys Acta. 1963 Feb 12;67:173–187. doi: 10.1016/0006-3002(63)91815-8. [DOI] [PubMed] [Google Scholar]
- Colby J., Dalton H., Whittenbury R. Biological and biochemical aspects of microbial growth on C1 compounds. Annu Rev Microbiol. 1979;33:481–517. doi: 10.1146/annurev.mi.33.100179.002405. [DOI] [PubMed] [Google Scholar]
- Colby J., Zatman L. J. Purification and properties of the trimethylamine dehydrogenase of bacterium 4B6. Biochem J. 1974 Dec;143(3):555–567. doi: 10.1042/bj1430555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J., Zatman L. J. Trimethylamine metabolism in obligate and facultative methylotrophs. Biochem J. 1973 Jan;132(1):101–112. doi: 10.1042/bj1320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUMMEL J. P., DREYER W. J. Measurement of protein-binding phenomena by gel filtration. Biochim Biophys Acta. 1962 Oct 8;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- Hartzell C. R., Beinert H. Oxido-reductive titrations of cytochrome c oxidase followed by EPR spectroscopy. Biochim Biophys Acta. 1976 Feb 16;423(2):323–338. doi: 10.1016/0005-2728(76)90189-4. [DOI] [PubMed] [Google Scholar]
- Hill C. L., Steenkamp D. J., Holm R. H., Singer T. P. Identification of the iron-sulfur center in trimethylamine dehydrogenase. Proc Natl Acad Sci U S A. 1977 Feb;74(2):547–551. doi: 10.1073/pnas.74.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large P. J., MacDougall H. An enzymic method for the microestimation of trimethylamine. Anal Biochem. 1975 Mar;64(1):304–310. doi: 10.1016/0003-2697(75)90435-2. [DOI] [PubMed] [Google Scholar]
- Orme-Johson W. H., Beinert H. Anaerobic reductive titrations with solid diluted sodium dithionite in an apparatus suitable for EPR spectroscopy. Anal Biochem. 1969 Dec;32(3):425–435. doi: 10.1016/s0003-2697(69)80010-2. [DOI] [PubMed] [Google Scholar]
- Steenkamp D. J., Beinert H. Mechanistic studies on the dehydrogenases of methylotrophic bacteria. 2. Kinetic studies on the intramolecular electron transfer in trimethylamine and dimethylamine dehydrogenase. Biochem J. 1982 Nov 1;207(2):241–252. doi: 10.1042/bj2070241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp D. J., Gallup M. The natural flavorprotein electron acceptor of trimethylamine dehydrogenase. J Biol Chem. 1978 Jun 25;253(12):4086–4089. [PubMed] [Google Scholar]
- Steenkamp D. J., Mallinson J. Trimethylamine dehydrogenase from a methylotrophic bacterium. I. Isolation and steady-state kinetics. Biochim Biophys Acta. 1976 May 13;429(3):705–719. doi: 10.1016/0005-2744(76)90319-3. [DOI] [PubMed] [Google Scholar]
- Steenkamp D. J., McIntire W., Kenney W. C. Structure of the covalently bound coenzyme of trimethylamine dehydrogenase. Evidence for a 6-substituted flavin. J Biol Chem. 1978 Apr 25;253(8):2818–2824. [PubMed] [Google Scholar]
- Steenkamp D. J., Singer T. P. Participation of the iron-sulphur cluster and of the covalently bound coenzyme of trimethylamine dehydrogenase in catalysis. Biochem J. 1978 Feb 1;169(2):361–369. doi: 10.1042/bj1690361. [DOI] [PMC free article] [PubMed] [Google Scholar]


