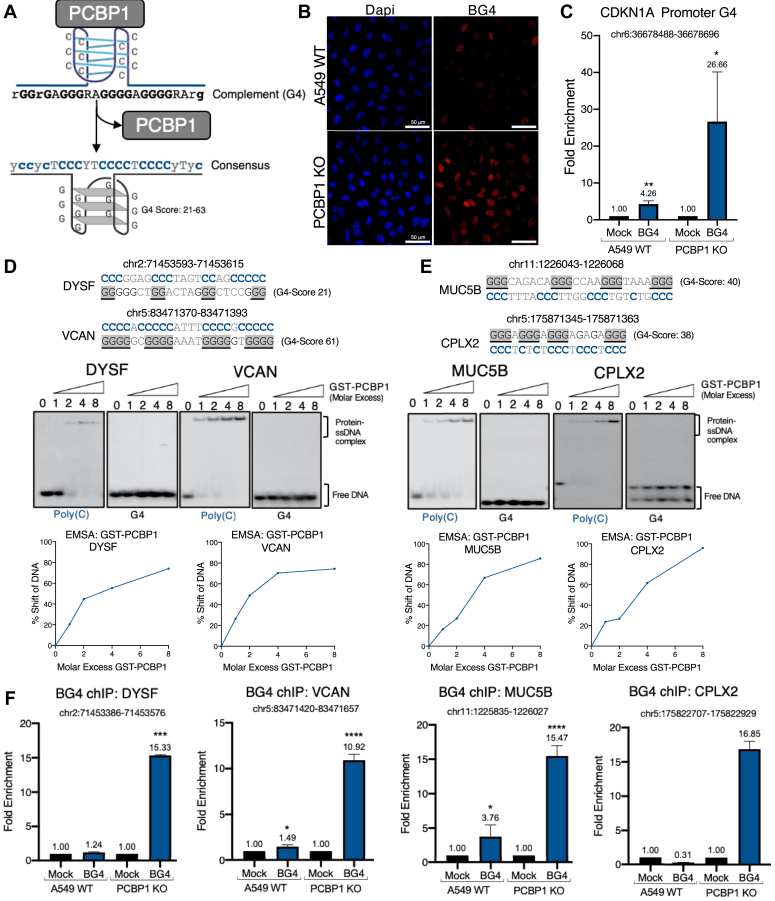
Figure 4.
Silencing PCBP1 leads to promoter G4 accumulation.A, model showing G4 formation upon PCBP1 depletion and QGRS score of G4-formed by sequence complementary to the PCBP1 consensus motif. B, immunofluorescence of A549 WT and PCBP1 KO cells stained with DAPI and BG4 (anti-G4 structure) antibody. C, BG4 ChIP-PCR of CDKN1A promoter DNA from A549 WT and PCBP1 KO nuclear lysates. D, GST-PCBP1 EMSAs of poly(C) sites in the promoters of upregulated differentially expressed genes, DYSF and VCAN. Representative graphs used to calculate 50% gel shifts for each probe are shown. E, GST-PCBP1 EMSAs of poly(C) sites in the promoters of downregulated differentially expressed genes, MUC5B and CPLX2. Representative graphs used to calculate 50% gel shifts for each probe are shown. F, BG4 ChIP-PCR of promoters of DYSF, VCAN, MUC5B, and CPLX2 from A549 WT and PCBP1 KO nuclear lysates. A single asterisk (∗) signifies a p-value less than 0.05, (∗∗) signifies p-value less than 0.01, (∗∗∗) signifies p-value less than 0.001, and (∗∗∗∗) signifies a p-value less than 0.0001.