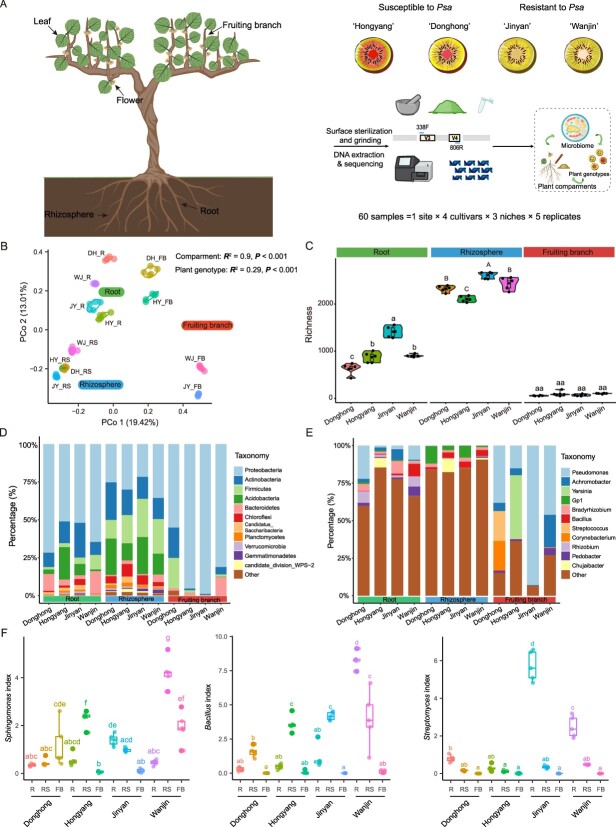
Figure 1.
Community structure and microbial diversity in three ecological niches of four kiwifruit cultivars. A Workflow for processing, DNA extraction, and 16S rRNA sequencing of 60 samples from different ecological niches of four kiwifruit cultivars in Qianshan, Anhui Province. B Unconstrained PCoA (PCo1 and PCo2) with Bray–Curtis distances showing that the kiwifruit (cvs ‘Hongyang’, ‘Donghong’, ‘Jinyan’, and ‘Wanjin’) microbiomes from different ecological niches (fruiting branch endosphere, root endosphere, and rhizosphere) were separated on the first axis. C Richness indices of fruiting branch-, root-, and rhizosphere-associated bacteria for four plant cultivars. D, E Relative abundance of the top 12 dominant phyla (D) and dominant genera (E) in three ecological niches of different kiwifruit cultivars. F Box plots showing that the abundances of three bacterial genera (Bacillus, Sphingomonas, and Streptomyces) in the three ecological niches of the disease-resistant cultivar ‘Wanjin’ were significantly greater than those in the disease-susceptible cultivars (‘Donghong’ and ‘Hongyang’). The different letters represent significant differences (P < 0.05). R, root; RS, rhizosphere; FB, fruiting branch; HY, Hongyang; DH, Donghong; JY, Jinyan, WJ, Wanjin.