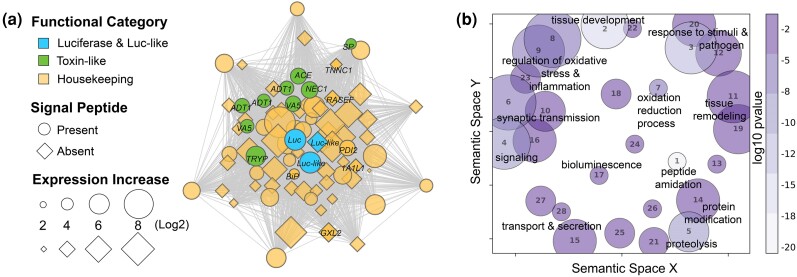
Fig. 2.
a) The BCN represents a group of co-expressed genes that are associated with the expression of luciferase, luciferase-like genes, toxin-like genes, and housekeeping genes some of which are constituents of stress and secretory protein pathways. The BCN comprises over 900 co-expressed genes and about one-third of the BCN genes are significantly upregulated uniquely in the upper lip of V. tsujii, of which 26% are considered integral to the network (MM > 0.8). Out of 277 co-expressed genes upregulated in the BUL, only genes with annotations (30%, 83 annotated genes) are plotted here to visualize connections and overall network topology. Bioluminescent genes, including c-luciferase (Luc) and similar but not functionally tested luciferase-like (Luc-like) genes are blue. Putative toxin-like genes are green: Venom Allergen 5 (VA5), A disintegrin and metalloproteinase with thrombospondin (ADT1), SP inhibitor (SP), trypsin-1 (TRYP), neuroendocrine convertase 1 (NEC1), and angiotensin-converting enzyme (ACE). Protein secretory pathway genes are in yellow with black labels: PDI2, BiP, RASEF, glucoside xylosyltransferase 2 (GXLT2), troponin C (TNNC1), and 1-aminocyclopropane-1-carboxylate synthase-like protein 1 (1A1L1). Genes that have a signal peptide and are presumably secreted outside the cell are indicated by a circle and genes that do not have a signal peptide are indicated by a diamond. Distance between nodes indicates strength connectivity. b) GO terms enriched in the BCN are visualized using GO figure. Each bubble represents a GO term or a cluster of GO terms summarized by a representative term reported in the supplement. The size of the bubble indicates the number of GO terms in each cluster and the color is the P-value or the average P-value of the representative GO term in that cluster. The GO terms for each bubble are found in supplementary table S3, Supplementary Material online.