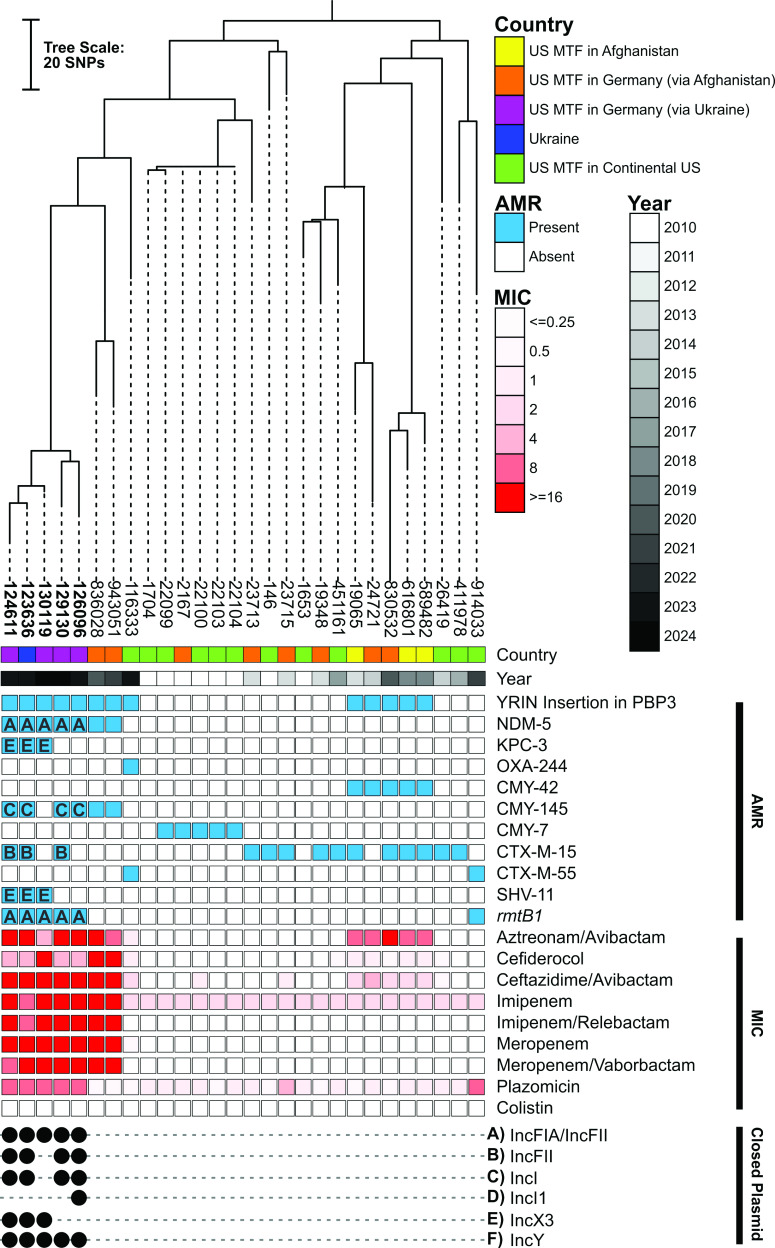
Fig 1.
A core genome phylogenetic tree of ST-361 E. coli (n = 28) from the US Military Health System and Ukraine. Country of origin, year of collection, and presence (closed square) or absence (open square) of selected antimicrobial resistance genes and minimum inhibitory concentrations of selected antibiotics. Replicon typing (A–F) of closed plasmids is indicated by the closed black circle. For closed sequences (label in bold font), the genetic location of antimicrobial resistance genes is indicated by the letter of the replicon type identified in the plasmid. The midpoint was used as a root for the phylogenetic tree.