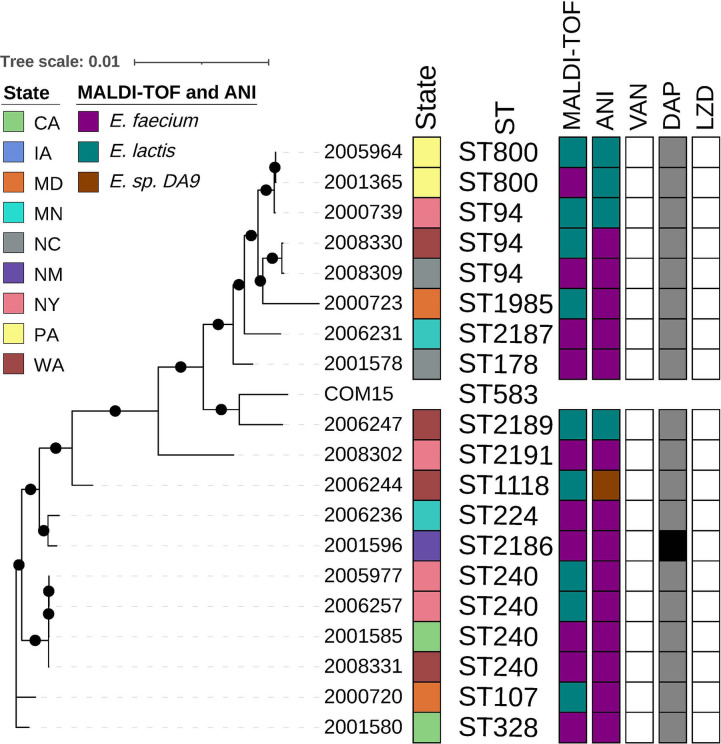
Fig 3.
E. lactis maximum likelihood phylogeny based on core gene alignments generated by Roary and RaxML. Annotation (from left to right) shows isolate ID, state of origin, multi-locus sequence type (ST), MALDI-TOF MS identified species, ANI identified species, minimum inhibitory concentration (MIC) interpretations for vancomycin, daptomycin, and linezolid. ST obtained using E. faecium scheme from PubMLST, MIC interpretations following E. faecium guidance represents susceptible in white; susceptible dose-dependent in gray; and resistant in black. Black circles on branches represent 100% support for the branch out of 100 bootstraps. Abbreviations: LZD, linezolid; DAP, daptomycin; VAN, vancomycin; ST, multi-locus sequence type; CA, California; IA, Iowa; MD, Maryland; MN, Minnesota; NM, New Mexico; NY, New York; NC, North Carolina; PA, Pennsylvania; and WA, Washington.