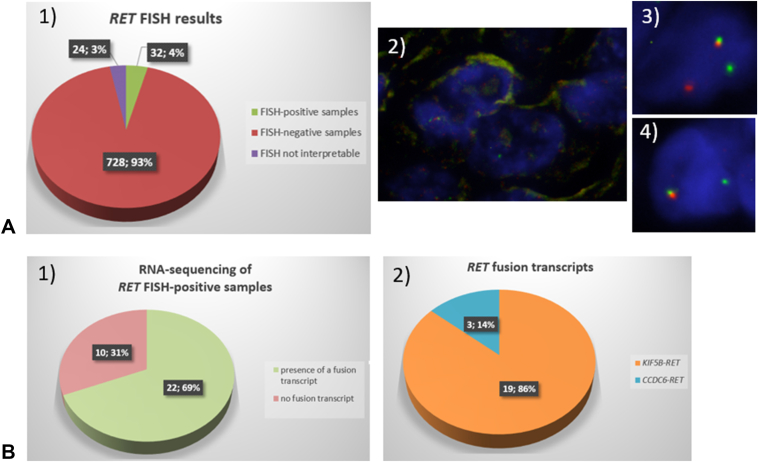
Figure 3.
RET-FISH results on the 784 NSCLC samples analyzed and targeted RNA-seq results on the 32 RET-FISH positive samples. (A) RET-FISH results on the 784 NSCLC samples of our NSCLC cohort. (1) Repartition of the RET-FISH positive, negative, and not interpretable samples. (2) Example of a non-interpretable result (signal intensity too low), (3) Example of a RET-FISH positive nucleus showing a split signal, (4) Example of a RET-FISH positive nucleus showing an isolated 3′ (green) signal. (B) Targeted RNA-seq results on the 32 RET-FISH positive samples. (1) Number and proportion of RET-FISH positive cases for which a fusion transcript was present or absent. (2) Nature and repartition of the fusion transcripts detected. FISH, fluorescence in situ hybridization.