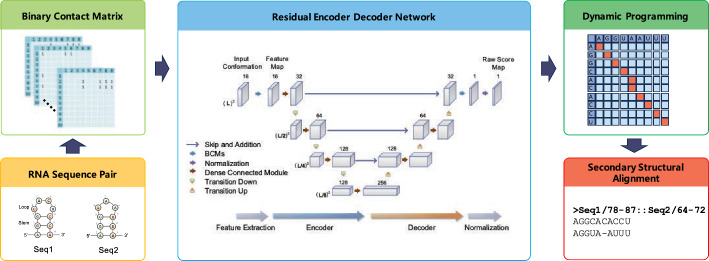
Fig. 1.
Graphical overview of REDalign architecture. REDalign includes three main steps to derive a precise RNA structural alignment. First, REDalign transforms the pair of input RNA sequences into two-dimensional binary contact matrices in order to represent the relative position of dinucleotides in the RNA sequences. Next, through the residual encoder and decoder network architecture, it learns the conserved structures of RNA sequences and can yield the alignment probability of dinucleotides within RNA sequences. Finally, REDalign derives the accurate structural alignment of RNA sequences using dynamic programming