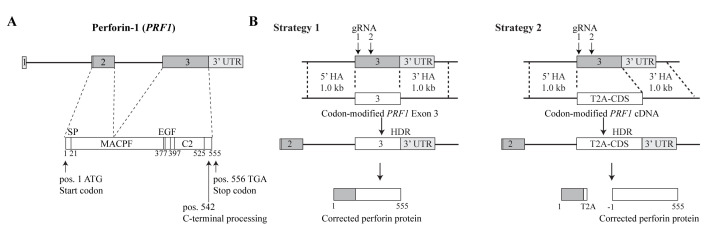
Figure 1. Examples of two gene correction strategies for the PRF1 gene.
A. Schematic representation of the human PRF1 gene and the encoded 555 amino acid perforin polypeptide. UTR, untranslated region; SP, signal peptide (cleaved off after translocation of the nascent protein into the ER); MACPF, membrane attack complex/perforin domain (includes the central machinery of pore formation); EGF, EGF-like domain (forms a shelf-like assembly connecting MACPF and C2 domain); C2, C2 domain (calcium-dependent phospholipid binding). B. Two gene correction strategies. With both strategies, the native exon 3 coding part gets replaced by a repaired version whose sequence has been diversified (codon-modified, see Procedure step B2). Repair templates are codon-modified to prevent unwanted HDR events that do not lead to gene repair. Strategy 1 replaces the mutated version of exon 3 with a repaired version. For a more comprehensive gene correction by cDNA knock-in [25], strategy 2 replaces exon 3 with the full-length PRF1 coding sequence (CDS): In place of the 5′ UTR, we engineered the cDNA to start with a T2A “self-cleaving peptide” preceded by a flexible serine-glycine-linker (TCC.GGC.AGC.GGC) [26], that is followed by the PRF1 ATG start codon, the signal peptide, and the complete PRF1 coding sequence ending with the TGA stop codon. Strategy 2 places the PRF1 CDS under endogenous transcriptional and translational control and allows correction of the ~60 known pathogenic PRF1 mutations (with the exception of frameshift or nonsense mutations in exon 2) [12,13,27]. Cleavage sites for the two guide RNAs (gRNA) are indicated. We use two gRNAs for gene repair in primary T cells because this increases the gene repair efficiency (see Li et al. [2] Suppl. Table S1). HA, homology arm; HDR; homology-directed repair. Adapted from Figure 6 in Li et al. [2].