Fig. 1 |. Alignments and constructs.
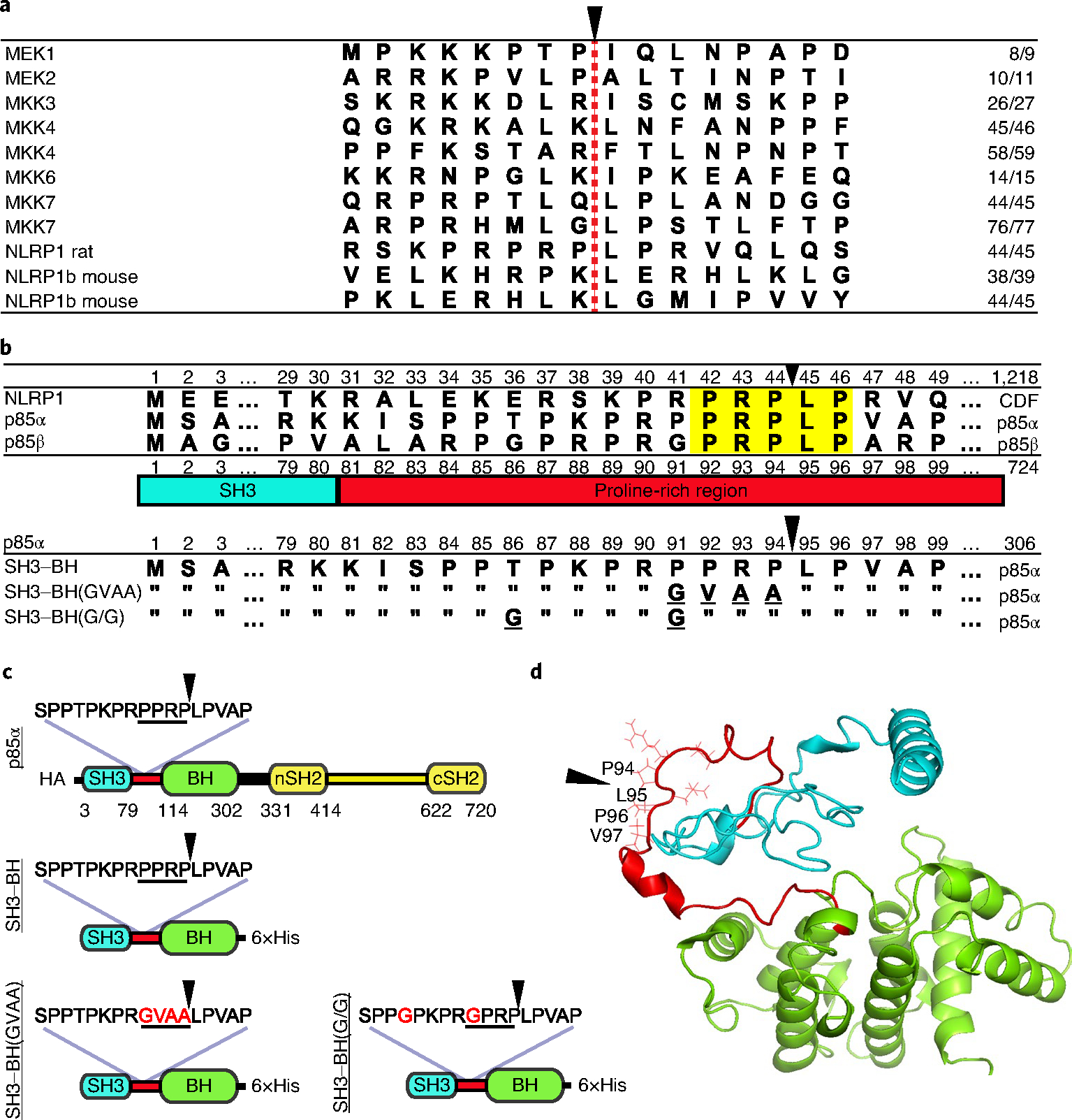
a, Alignment of cleavage regions of LF substrates. The LF cleavage site is denoted with a red dashed line and a black arrowhead, and the residue number of the cleavage site is listed on the right. b, Alignment of N-terminal amino acid sequences of CDF (Fischer) rat strain NLRP1 with human p85α and p85β. Highlighted in yellow are the five amino acids in the p85 proteins matching the P3–P2′ positions of the rat NLRP1 LF cleavage site, which is indicated with a black arrowhead. The PI3K p85 SH3 domain (blue) and proline-rich linker region (red) are shown below the alignment. Bottom alignments show p85α constructs used in this paper, which include a truncated variant (SH3–BH) in which residues 1–306 are expressed, along with mutated variants. Underlined letters indicated mutated residues, and residues identical in the mutated constructs are indicated with double quotes. c, Domain layout of full-length and truncated constructs listed in b with amino acid substitutions in red font. HA, N-terminal 3×haemagglutinin tag. d, Predicted structure (model by iTASSeR) of the SH3 domain, proline-rich linker region and BH domain of p85α; C-score of −0.9 (refs. 10–12). Black arrowheads in c and d indicate the LF cleavage site.
