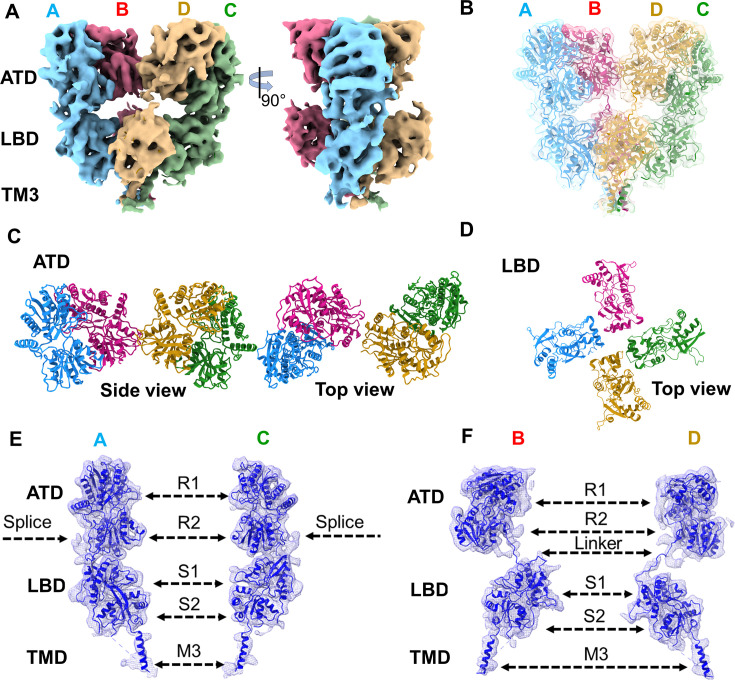
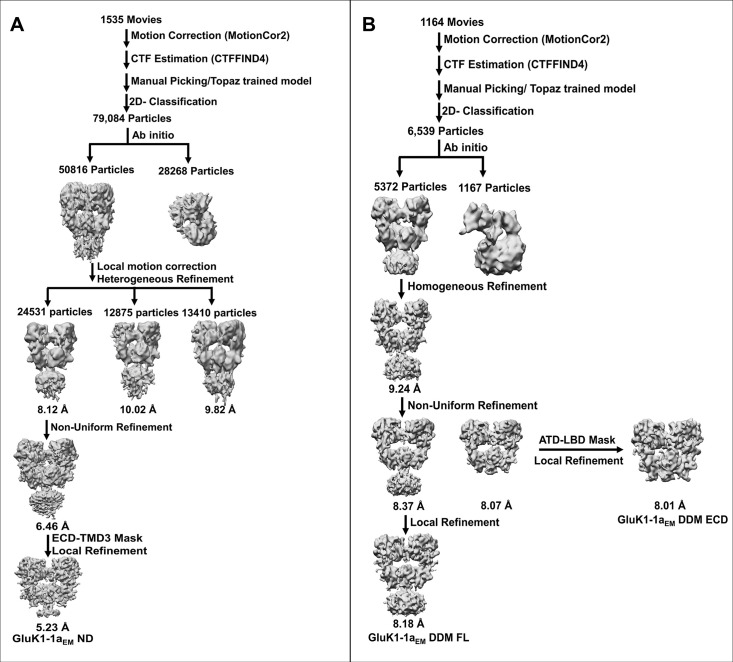
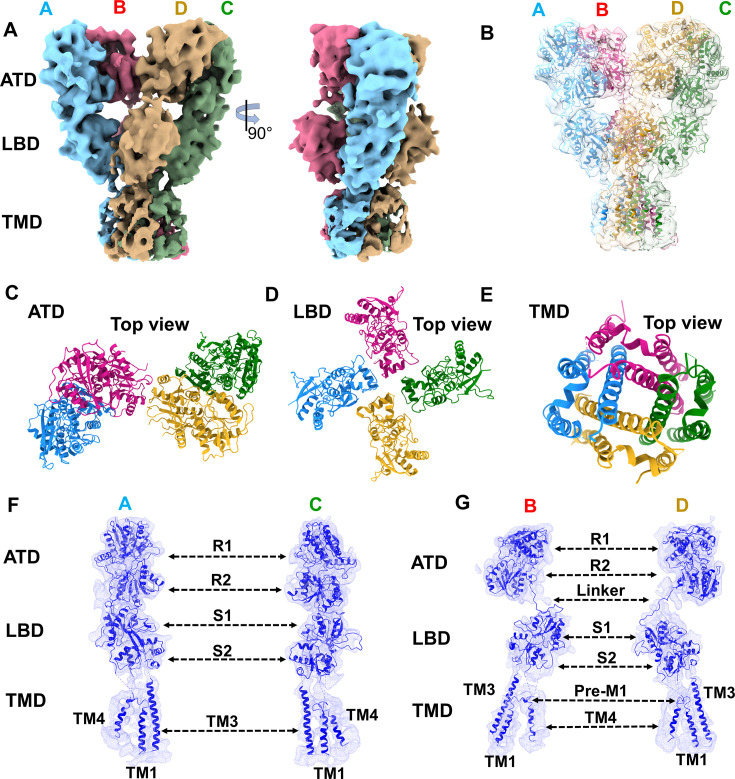
Figure 6. Architecture of GluK1-1aEM reconstituted in nanodisc for SYM-bound desensitized state.
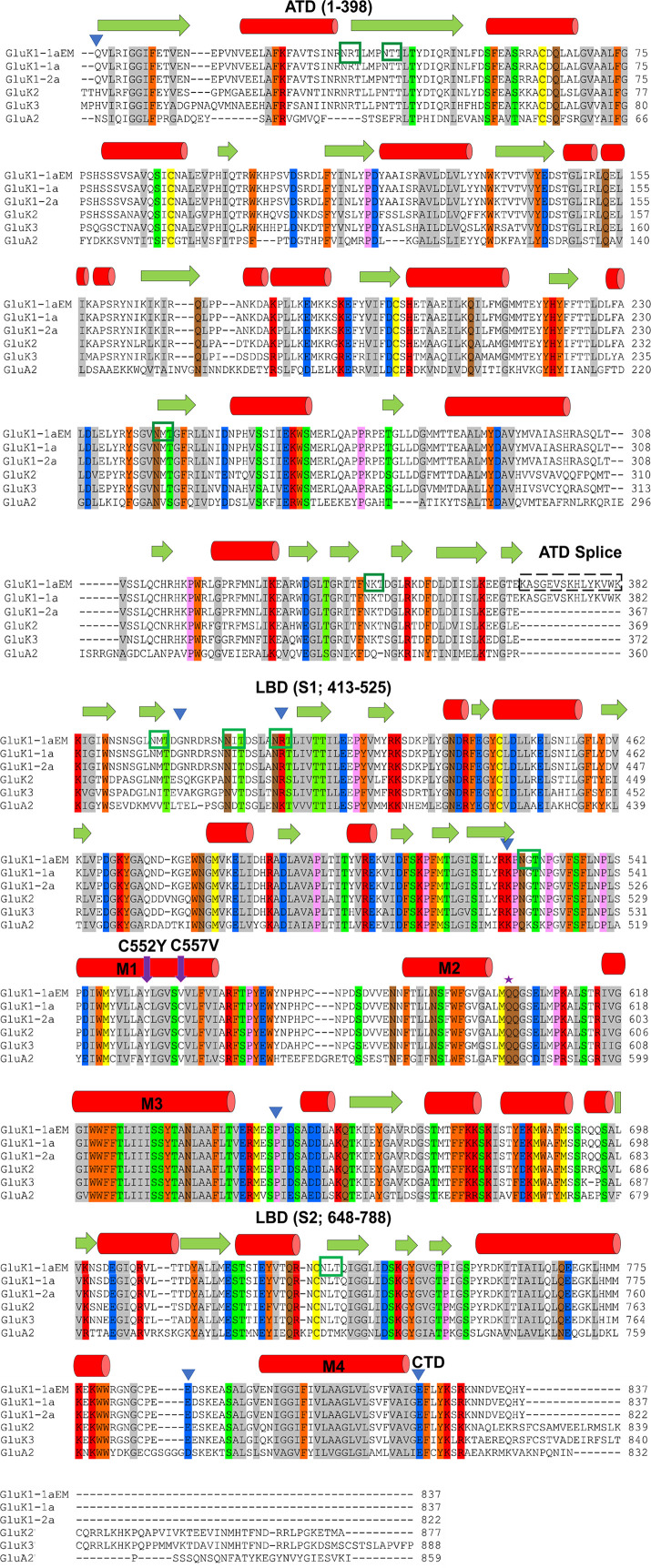
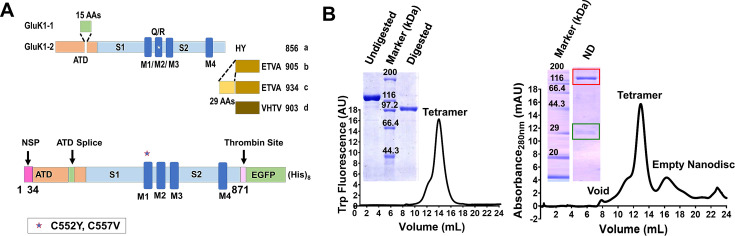
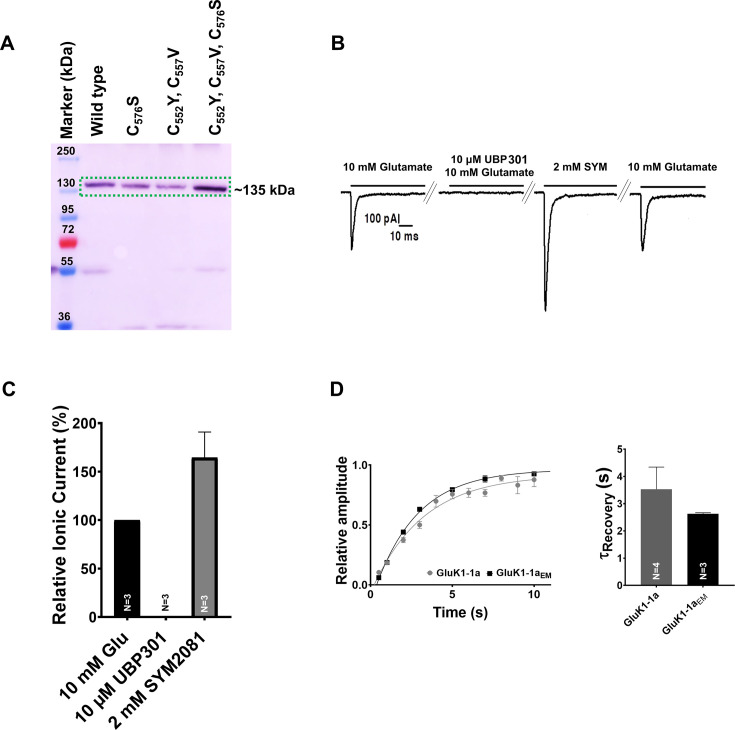
(A) Shows the segmented density map colored according to unique chains of the receptor tetramer (A- blue, B-pink, C-green, and D-gold) at 5.23 Å in side view and 90° rotated orientations. (B) Shows the final model fitted in the EM map. (C and D) Top views of amino-terminal domain (ATD) and ligand binding domain (LBD) layers. (E & F) Display the segmented map fitted with the corresponding distal (A & C) and proximal (B & D) chains. Receptor sub-domains, the position of splice insertion, and linkers are indicated.