Fig. 2.
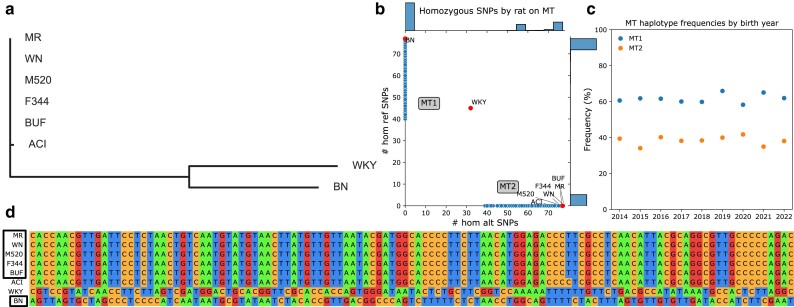
MT haplotypes present in HS founders and modern HS rats. a) NJ, unrooted tree using MT SNPs and indels in HS founders. Branch lengths correspond to genetic distance. b) Distribution of alleles by rat among MT SNPs passing filters (Supplementary Fig. S2). Plot shows count of reference alleles on X-axis and count of alternate alleles on Y-axis for each rat. Side plots are histograms of allele counts in modern HS rats (small dots). Missingness in low-coverage modern samples leads to scatter on the axes. Labeled large dots are HS founders. MT1 and MT2 haplogroups are labeled. c) Distribution of MT haplotypes in the HS rat population over time. Plot shows birth year on X-axis and haplotype percentage on Y-axis. d) Pseudo-alignment of founder haplotypes, at all MT SNPs called by low-coverage sequencing, colored by nucleotide. Boxed founders have haplotypes present in modern HS rats.