Figure 1.
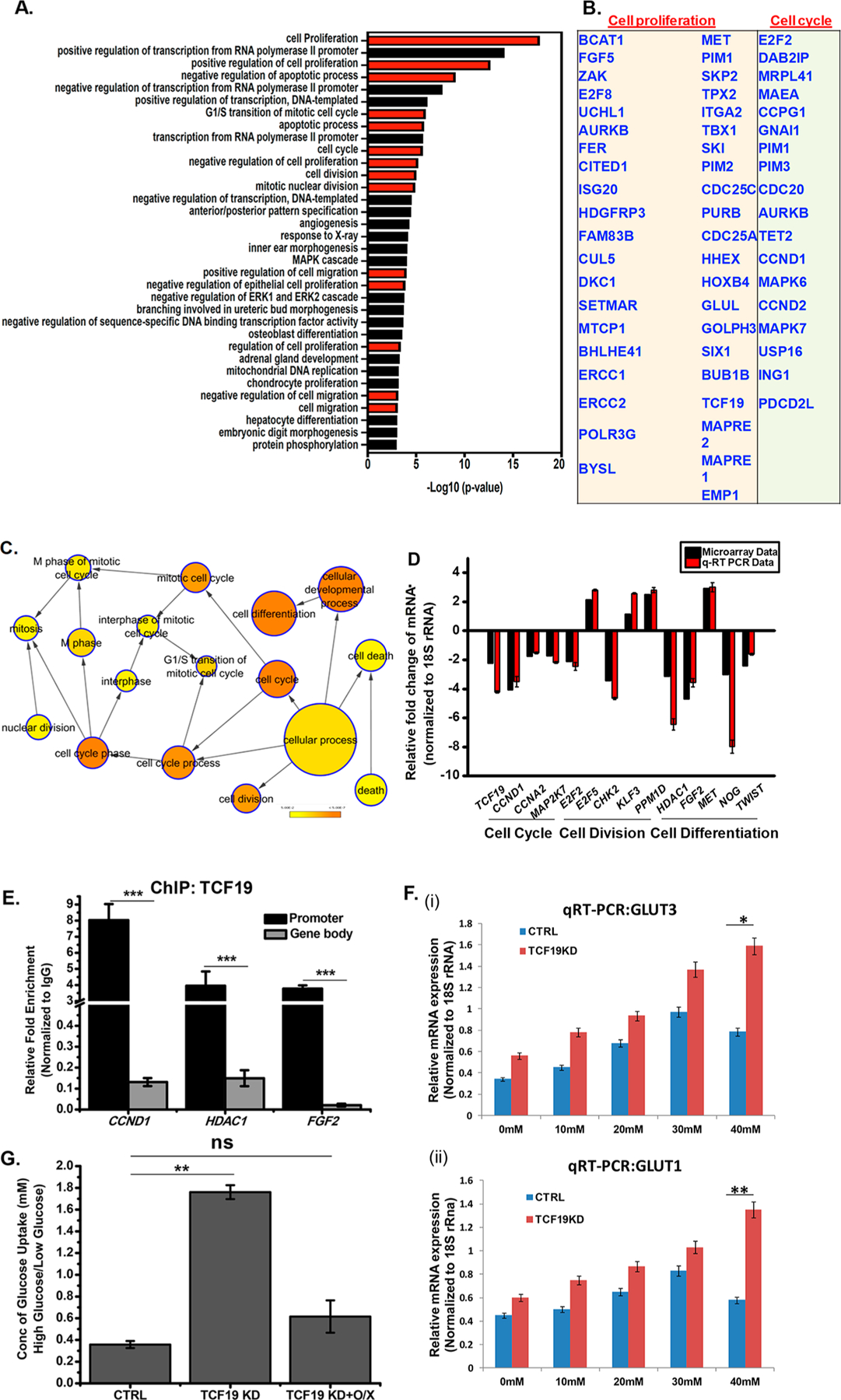
TCF19 is an important player in cellular proliferation. (A) Gene ontology (biological process) categories showing the most significant enrichment in a list of proliferation-associated genes obtained after microarray analysis upon TCF19 knockdown. GO terms are indicated on the Y-axis. The −log 10 P value on the X-axis indicates the level of significance of each process as obtained from the DAVID tool. (B) List of key cell proliferation- and cell cycle-related genes. (C) Genes regulating cellular proliferation that were differentially expressed at HG after knocking down TCF19 were analyzed by BiNGO. Fold change 2.0; p ≤ 0.05. (D) Validation of microarray genes for proliferation was performed by knocking down TCF19 under high-glucose conditions. (E) TCF19 is recruited to the promoter of the proliferation genes, but not on the gene body as observed by the ChIP assay. (F) qRT-PCR analysis of GLUT3 and GLUT1 in the presence and absence of TCF19 with an increasing glucose concentration. The presence of TCF19 represses the overexpression of GLUT receptors. (G) Glucose uptake assay to measure the cellular glucose uptake ability in the presence and absence of TCF19 under low- and high-glucose conditions. TCF19 KD stable cells were complemented with the FL FLAG-TCF19 construct that compensated for the cellular glucose uptake. Cells were glucose starved for 6 h followed by treatment with 10 μM 2-NBDG for 2 h.
