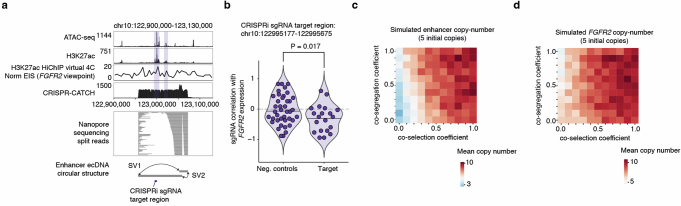
Extended Data Fig. 9. Structure and dynamics of the enhancer ecDNA.
(a) From top to bottom: ATAC-seq, H3K27ac ChIP-seq, H3K27ac HiChIP contact with the FGFR2 promoter, CRISPR-CATCH sequencing of enhancer-only ecDNA species in SNU16 cells, individual split reads in Nanopore sequencing supporting the circular enhancer-only ecDNA species, and structural variants (SV1 and SV2) that create a circular structure. Enhancer region targeted by CRISPR interference single-guide RNAs (sgRNAs) is marked at the bottom. SV1: precise inversion between chr10:122957191 and chr10:123051954; SV2: precise inversion between chr10:123058196 and chr10:123071737. (b) Correlations between individual sgRNAs and FGFR2 expression after CRISPR interference followed by sorting of cells with various levels of FGFR2 expression (data published and described in Hung et al.4). P-values determined by lower-tailed t-test compared to negative controls. Each dot represents an independent sgRNA (n = 40 negative control sgRNAs, n = 20 target sgRNAs). (c-d) Simulated copy number of enhancer-only ecDNA (c) and FGFR2 ecDNA (d) under various settings of co-selection and co-segregation. Individual selection on the enhancer-only species was kept at 0.0, and individual selection on the FGFR2 ecDNA was kept at 0.2. One million cells were simulated from a parent cell carrying 5 copies of both species. 10 replicates were simulated and the average value was reported. Norm EIS, normalized enhancer interaction signal.