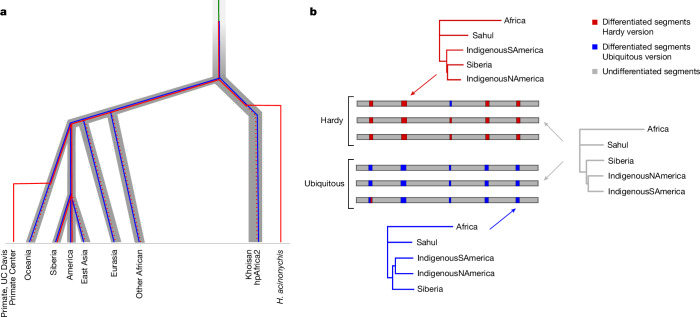
Fig. 2. Divergence and spread of Hardy and Ubiquitous gene pools.
a, Hypothesized scenario for the differentiation of H. pylori into two ecospecies and subsequent global spread (not to scale). Thick grey tree represents a simplified history of human population differentiation (based on ref. 49). Helicobacter evolution is represented by thinner lines, which are within the grey tree during periods of evolution in humans. Green line represents H. pylori before the evolution of the two ecospecies; blue line represents Ubiquitous ecospecies; red line represents Hardy ecospecies; dotted red lines indicate that Hardy strains have not yet been detected on the branch and therefore may have gone extinct. b, Phylogenetic population trees for differentiated (Hardy in red and Ubiquitous in blue) and undifferentiated (in grey) regions of the genomes. Trees were constructed considering only populations with both Hardy and Ubiquitous representatives. Pairwise distances between strains were calculated for the relevant genome regions, and population distances were calculated by averaging over pairwise strain distances. For Africa, we used H. acinonychis strains for the Hardy differentiated gene trees and hpAfrica2 strains for the Ubiquitous differentiated gene tree. For the undifferentiated gene tree, we averaged over the relevant Hardy and Ubiquitous strains.