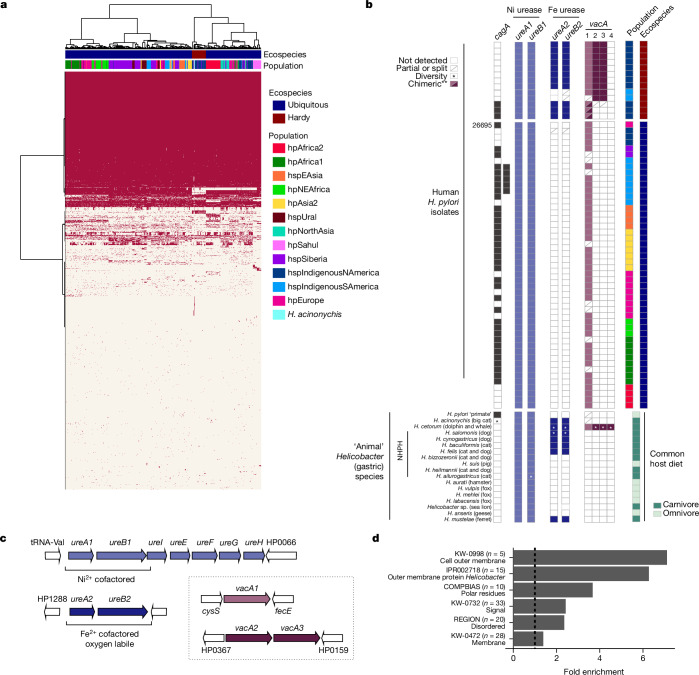
Fig. 4. Genome composition in human and animal Helicobacter.
a, Hierarchical clustering based on pangenome presence (red)/absence (white) for a sample of the global dataset. Strains are coloured based on their population and ecospecies. b, Top, presence/absence of cagA and different vacA and ureA/B types in Hardy and Ubiquitous H. pylori from humans. Included is a subset of complete genomes, two cagA+ Hardy strains and reference strain 26695. Bottom, presence/absence in gastric Helicobacter isolated from animals. Common host(s) and their diets are indicated (Supplementary Table 4). c, Representative configuration and genomic context of ureA/B and vacA in Hardy H. pylori genomes, based on strain HpGP-CAN-006. Lighter-coloured arrows indicate genes present in both ecospecies, based on sequence and genomic context/synteny; darker-coloured arrows indicate Hardy-specific versions. d, Fold enrichment for significant (P < 0.05, one-sided Fisher’s test, Benjamini correction) functional terms. Chimeric, potential chimeric version (Hardy + Ubiquitous); Diversity, differential presence/absence in analysed genomes within the species; NHPH. non-H. pylori Helicobacter spp.; tRNA, transfer RNA.