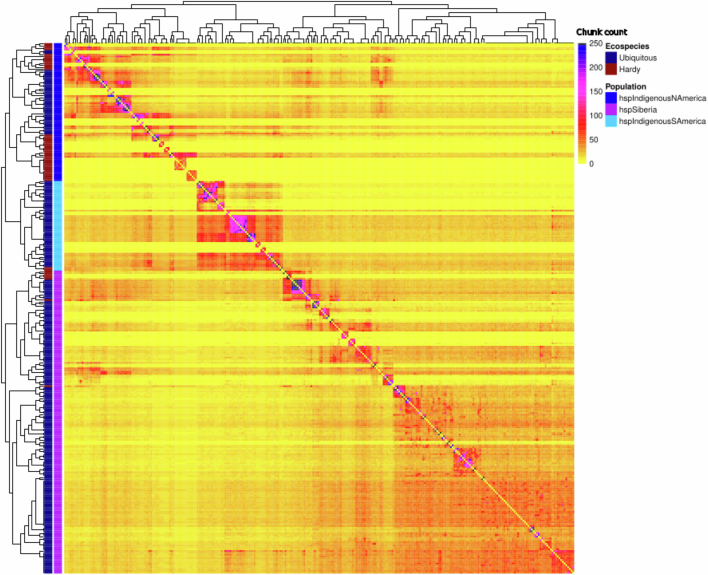
Extended Data Fig. 4. FineSTRUCTURE analysis of the strains from hspSiberia, hspIndigenousNAmerica and hspIndigenousSAmerica.
The strains from the Hardy clade are highlighted by red shading, overlaying the dendrogram on the left of the plot, while the Ubiquitous strains are highlighted with blue. FineSTRUCTURE uses an in silico chromosome painting algorithm to fit each strain as a mosaic of nearest neighbours, chosen from the other strains in the dataset. Each row shows the coancestry vector for one strain, which is a count of the number of segments of DNA used in the painting from each of the other strains in the dataset. High coancestry between strains implies that they are nearest neighbours for many segments of the genome and hence share genetic material from a common gene pool. FineSTRUCTURE based clustering is more sensitive to recent gene flow than clustering using genetic distances.