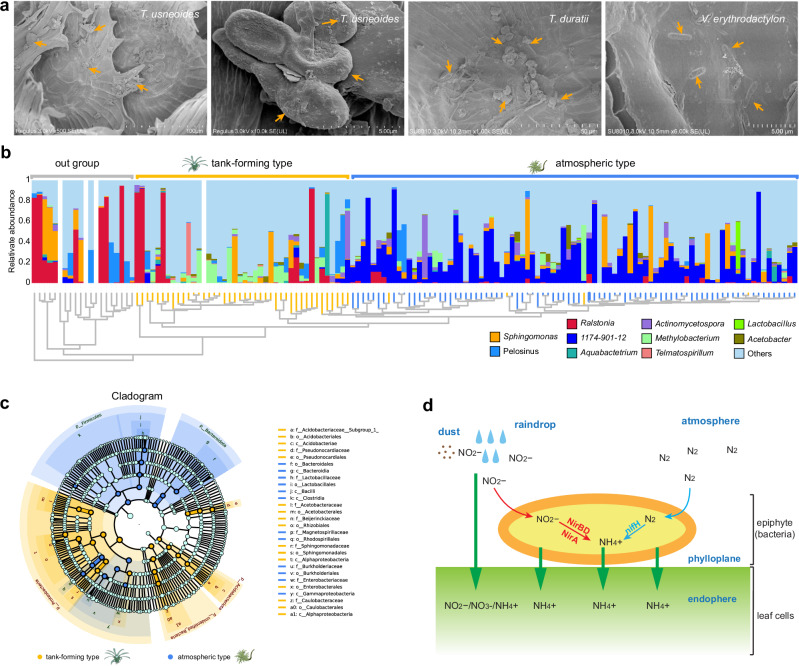
Fig. 7. Identification and phylogeny of nitrogen-fixing bacteria in the tillandsioid phyllosphere.
a SEM revealing microorganisms on the leaf surfaces of T. duratii and V. erythrodactylon. The orange arrows indicate bacteria or their excretions. b Top-10 phyllospheric bacterial communities of 127 tillandsioids and 17 out-group bromeliads with genera divergence. The phylogenetic tree constructed by bacterial communities mirrors the phylogenetic tree based on tillandsioids genera divergence as shown in Fig. 1. The relative abundance of phyllospheric bacterial communities is presented at the genus level, with different genera depicted in different colors. c Cladogram generated from the LEfSe analysis indicating the phylogenetic distribution from phylum to genus of the microbiota of two types of tillandsioids. In the phylogenetic tree, the concentric circles radiating outward represent taxonomic levels from phylum to genus (or species). Each small circle at different taxonomic levels represents a classification at that level, with the diameter of each circle proportional to its relative abundance. Species with no significant differences are uniformly colored light blue. Species (or genera, or families, or orders, or phylum) with differences are colored according to their groups; orange nodes represent bacterial groups that play important roles in tan-forming plants, while dark blue nodes represent those in atmospheric-type plants. Species (or genera, or families, or orders, or phylum) names represented by letters in the figure are explained in the legend at the bottom. d Nitrogen metabolism pathways in T. usneoides phyllospheric bacterial communities. Two ammonification pathways were identified in the phyllospheric bacterial metagenome, including the N2-fixation and Nitrate reduction pathways.