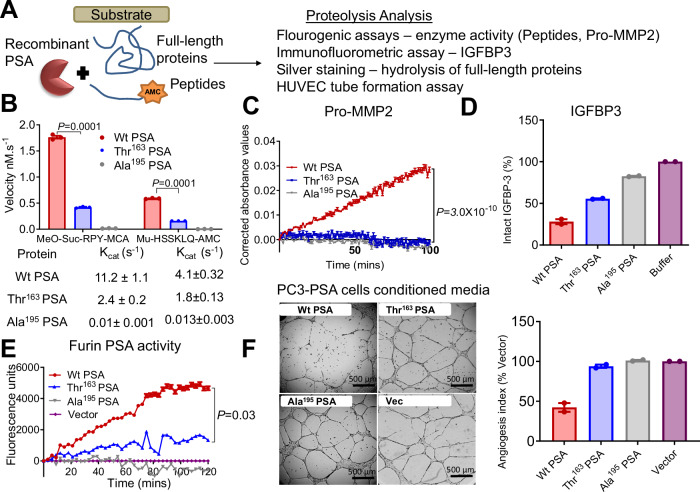
Fig. 3. Biochemical characterisation of the effect of the Thr163 variant on PSA activity.
A Schematic for PSA proteolytic activity analysis. B Rate of hydrolysis by mature PSA proteins (Wt PSA, Thr163 PSA, and catalytically inactive mutant control Ala195 PSA, all at 0.1 µM) were compared using the peptide substrates MeO-Suc-RPY-MCA (10 µM) and Mu-HSSKLQ-AMC (1 µM) over 4 h at 37 °C. Proteolytic activity derived from assaying a constant amount of PSA with increasing concentration (0–250 mM) for these two substrates were used to estimate Kcat values using nonlinear regression analysis in Graphpad Prism. (n = 3 independent experiments). Also see Supplementary Fig. 5B. C Time (mins) versus relative absorbance (OD) corrected to the substrate alone controls was plotted indicating the activity of pro-MMP2 (0.14 µM) when pre-incubated with PSA protein variants (Wt, Thr163 and Ala195 at 0.07 µM) and then the activity analysed with the chromogenic substrate (Ac-PLG-[2-mercapto-4-methyl-pentanoyl]-LG-OC2H5, 40 µM) for active MMP2 over 2 h. (n = 3 independent experiments). D Intact/total IGFBP-3 (2.2 µM) after 24 h incubation with PSA variants (0.25 µM) as shown relative to IGFBP-3 control without added PSA. Also see Supplementary Fig. 5C. (n = 2 independent experiments). E Fluorescent activity observed for the furin generated active PSA captured from serum free conditioned media of furin-PSA overexpressing PC-3 cells (Wt, Thr163, inactive mutant Ala195 and vector) using the peptide substrate MeO-Suc-RPY-MCA (n = 3 independent experiments). F Inhibition of HUVEC tube formation on Matrigel by treatment of HUVECs with serum free conditioned media from the PC-3 cells overexpressing (Wt, Thr163 and Ala195 PSA) and Vector control. Scale bar is 500 µm. The graph to the right represents the effect of these PSA protein variants on HUVEC tube formation expressed as an angiogenesis index49,65 is shown in relation to the control (n = 2 independent experiments). This is complemented by the same assay using recombinant PSA. Also see Supplementary Fig. 5D. All error bars represent mean ± SEM. Statistical analyses were determined by two-sided Student’s t test (B) or one-way ANOVA followed by Dunn’s multiple comparison test (C, E). Source data are provided as a Source Data file.