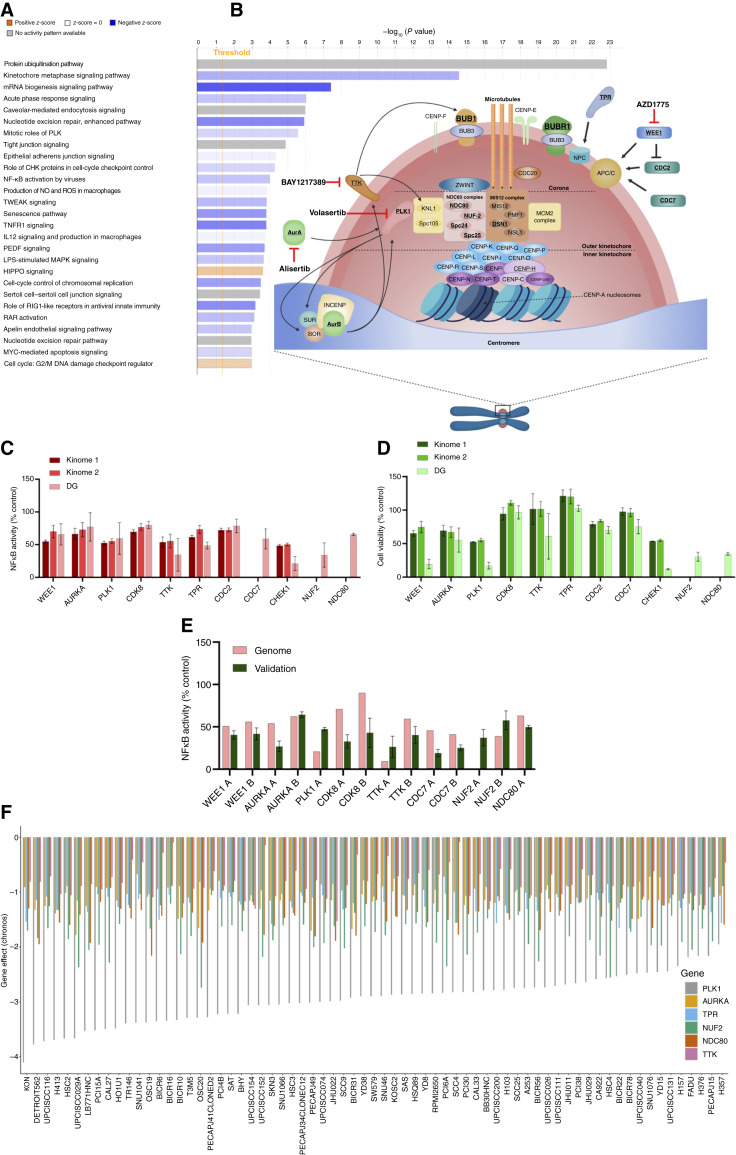
Figure 2.
Identification of G2/M checkpoint and kinetochore components as modulators of TNF-induced NF-κB activity in HNSCC cells. A, IPA was performed using data for the 769 candidates for which depletion significantly modulating NF-κB activity and ranked as < −2 LogP reporter seed corrected by CSA (Supplementary Table S2). B, Schematic demonstrating identified G2/M checkpoint kinases and kinetochore components significantly modulating NF-κB activity in UMSCC1κB cells. Identified hits are in bold and underlined. C, NF-κB reporter activity in UMSCC1κB after transfection with select siRNAs from the kinome 1, kinome 2, and DG RNAi screens. NF-κB activity is shown as a percentage of a negative control siRNA (set at 100%). D, Cell viability in UMSCC1κB after transfection with select siRNAs from the kinome 1, kinome 2, and DG RNAi screens. Cell viability is shown as a percentage of a negative control siRNA (set at 100%). E, Validation of NF-κB reporter activity in UMSCC1κB after transfection with independent siRNAs. Select siRNAs from the secondary RNAi screen are shown compared with the DG results (genome in figure). NF-κB activity is shown as a percentage of a negative control (set at 100%). Each RNAi screen contained three individual siRNA targeting each gene. Validation siRNA experiments used separate siRNAs not used in the RNAi screens. F, Chronos-inferred gene dependency scores for PLK1, AURKA, NUF2, NDC80, and TTK in 69 head and neck cancer cell lines in the CRISPR knockout screens from Broad’s Achilles and Sanger’s Score projects. The order is based on the gene effect of PLK1 [most dependent cell line (lowest value) to least dependent cell line (highest value); B, created with BioRender.com.]