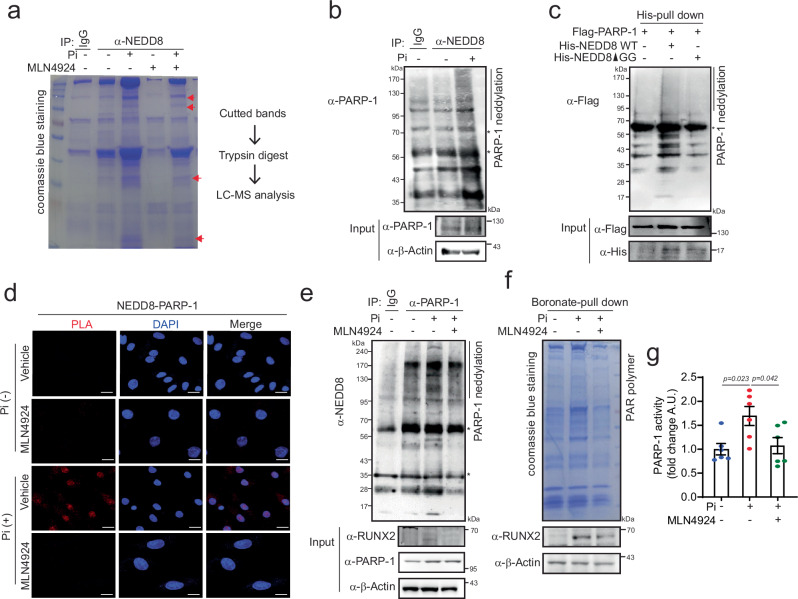
Fig. 2. NEDD8 is conjugated to PARP-1 in VC.
a A gel stained with Coomassie blue for mass spectrometry analysis. Cell extracts from A10 cells treated with Pi (4 mM) or MLN4924 (1 μM) were immunopurified with an anti-NEDD8 antibody and subjected to SDS‒PAGE. The bands indicated by arrows were then digested with trypsin and analyzed by mass spectrometry. b Cell lysates from Pi-treated A10 cells were immunoprecipitated with anti-NEDD8 and immobilized with anti-PARP-1. PARP-1 neddylation increased in Pi-induced VC. c Whole-cell lysates from 293 T cells transfected with the indicated constructs were subjected to Ni-NTA pull-down. PARP-1 neddylation was blunted in NEDD8-ΔGG compared with that in NEDD8 WT. d A PLA was performed. NEDD8-conjugated PARP-1 is dissociated by MLN4924 (1 μM). Scale bar=20 μM. e Immunoprecipitation with anti-PARP-1. Pi-induced PARP-1 neddylation is attenuated by MLN4924 (1 μM) in A10 cells. PARP-1 activity was measured by detecting the poly(ADP)-ribosylation (PAR) polymer (f) and conducting a PARP-1 enzyme activity assay (g). Pi-induced PARP-1 activity was blunted by MLN4924 treatment in A10 cells. n = 6 per group, independent experiments. Note: “*” indicates a nonspecific band. A.U., arbitrary units. The data are presented as the means ± SEMs. Statistical significance was tested via ANOVA with Tukey’s HSD test.