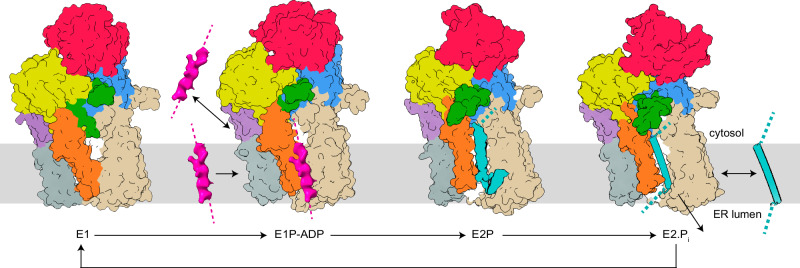
Fig. 6. Proposed polypeptide translocation mechanisms of P5A-ATPases.
The domains are colored coded as in Fig. 1b. P5A-ATPases seemingly rest in a cytosol-facing E1 state, which is blocked by the Plug domain (green). Autophosphorylation leads to displacement of the Plug-domain, and hence the cytosol-facing cavity becomes accessible to the cytosol in the E1P–ADP/E1P conformations. The unknown features (pink) in the latter structures may represent cargo (1) already removed from the ER membrane (in E1P, not shown in the model), and about to be bound for a new dislocation-cycle, or (2) signal-peptide, preparing the following peptide-stretch for membrane insertion. Polypeptide binding (cyan) in the all-through cleft is achieved in the E2P and E2.Pi states. The clearer secondary structure in E2.Pi may be (1) coincidental, or (2) an indication of folding of the inserted helix. Completion of the cycle, dephosphorylation, and reestablishment of the E1 state is associated with the release of the helix to (1) the cytosol as a removed helix or (2) the surrounding membrane or ER lumen as an inserted or secreted helix.