Fig. 1. Design strategy.
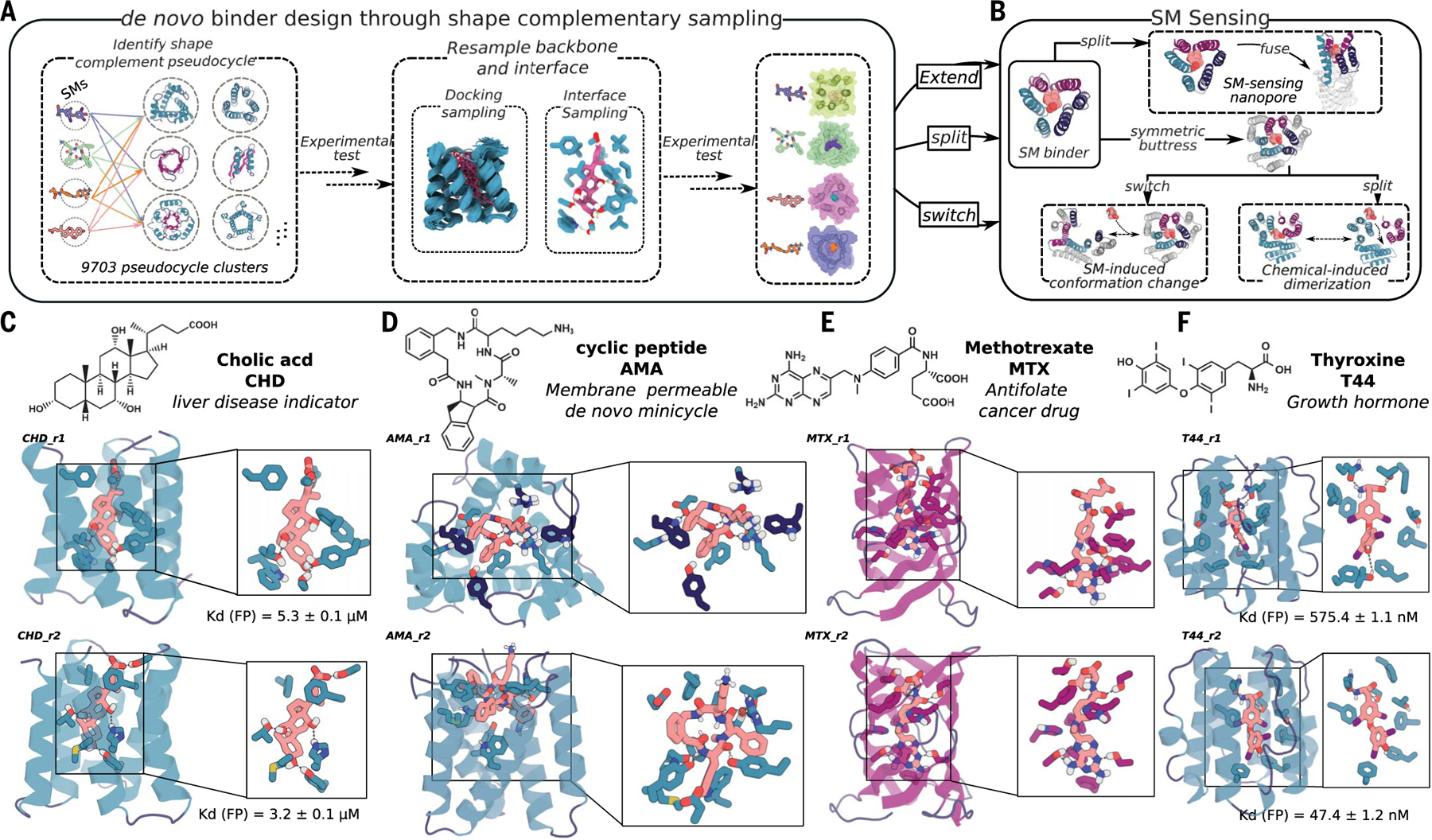
(A) Diverse conformers of the small molecule of interest are docked into deep learning–generated pseudocycles containing a wide array of central pockets, and the interface sequence is optimized for high-affinity binding with Rosetta or LigandMPNN. Top-ranked designs are tested experimentally, and the backbones of the best hits and the docked poses are extensively resampled. After sequence design, top-ranked second-round designs are experimentally tested (fig. S1, A and B). (B) Because pseudocycles are constructed from modular repeating units that surround the central binding pocket, the binders can be readily transformed into sensors through multiple strategies. (C to F) Examples of first-round design models for each target ligand.
