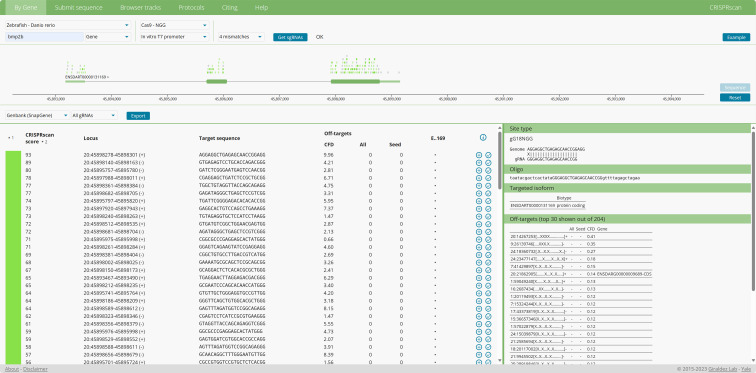
Figure 2. Example output from CRISPRScan.
Zebrafish-Danio rerio, Cas9-NGG, Gene, in vitro T7 promoter, and 4 mismatches should be chosen, and the gene symbol is then submitted to “Get sgRNAs”. This protocol uses Cas9 for genome editing, so we select Cas9-NGG to specify the prediction of sgRNA for Cas9. Allowing for “4 mismatches” enables the sgRNA site to tolerate up to four mismatches in off-target sequences. This criterion helps to identify a broader range of potential target sites for consideration.