Abstract
The building blocks-based molecular network (BBMN) strategy was applied to the phytochemical investigation of Cleistocalyx operculatus, leading to the targeted isolation of eighteen novel cinnamoylphloroglucinol-terpene adducts (CPTAs) with diverse skeleton types (cleistoperones A–R, 1–18). Their structures including absolute configurations were determined by extensive spectroscopic methods, quantum chemical calculations, and single-crystal X-ray crystallographic experiments. Cleistoperone A (1), consisting of a cinnamoylphloroglucinol motif and two linear monoterpene moieties, represents an unprecedented macrocyclic CPTA, whose densely functionalized tricyclo[15.3.1.03,8]heneicosane bridge ring skeleton contains an enolizable β,β′-triketone system and two different kinds of stereogenic elements (including five point and three planar chiralities). Cleistoperones B and C (2 and 3) are two new skeletal CPTAs with an unusual coupling pattern between the (nor)monoterpene moiety and the cinnamoyl chain of the cinnamoylphloroglucinol unit. Cleistoperone D (4) possesses an unprecedented cage-like 6/6/6/4/6-fused heteropentacyclic scaffold. The plausible biosynthetic pathways for 1–18 were also proposed. Notably, compounds 1, 4, 7, 8, and 18 exhibited significant antiviral activity against respiratory syncytial virus (RSV). The most potent one, cleistoperone A (1) with IC50 value of 1.71 ± 0.61 μmol/L, could effectively inhibit virus replication via affecting the Akt/mTOR/p70S6K signaling pathway.
Key words: Cleistocalyx operculatus, Cinnamoylphloroglucinol-terpene adduct, Targeted isolation, Molecular networking, Structure elucidation, Cleistoperone A, Respiratory syncytial virus, Antiviral activity
Graphical abstract
The BBMN strategy led to the isolation of eighteen unprecedented cinnamoylphloroglucinol-terpene adducts from Cleistocalyx operculatus. Cleistoperone A (1) showed potent anti-RSV activity via affecting the Akt/mTOR/p70S6K signaling pathway.
1. Introduction
Mass spectrometry (MS)-based metabolomics combined with other cheminformatics techniques have emerged as an efficient tool for natural product discovery1, 2, 3. Among them, tandem MS2 molecular networking can categorize massive amounts of MS2 fragmented ions into specific clusters on account of structural similarity, which has been widely used to dereplicate known compounds and prioritize the target compounds from complex crude extracts4, 5, 6. Particularly, the molecular networking is available on the Global Natural Products Social Molecular Networking (GNPS; http://gnps.ucsd.edu) open-access Web-based platform. By using this approach, a considerable number of analogues of the known compounds had been identified7, 8, 9, 10. However, the targeted discovery of novel natural products with significantly distinguishing skeletons from the known ones by classic molecular network is still a challenging task. Recently, by recognizing the characteristic MS2 fragment ions derived from the biogenetic building blocks of target compounds, our group developed a building blocks-based molecular network (BBMN) strategy for efficient discovery of biogenetically-related natural products with novel skeletons11, 12, 13. As exemplified by the successful identification of novel Securinega alkaloids with neuronal differentiation activities from the medicinal plant Flueggea suffruticosa, the BBMN strategy shows strong potential for the efficient discovery of unprecedented natural products that could afford new pharmacophoric scaffolds.
Respiratory syncytial virus (RSV) is the major pathogen of seasonal and global acute lower respiratory infection. Worldwide, RSV disease is estimated to cause more than 3.4 million hospitalizations and approximately 160,000 deaths in young children each year14. In infants less than one year of age, RSV is associated with significantly more deaths than influenza15. Despite the large medical and economic burden, treatment options available for RSV infection are limited. Ribavirin, palivizumab, and nirsevimab are licensed drugs approved for the treatment or prevention of RSV infection. Among them, ribavirin is a nucleoside analogue with broad-spectrum antiviral activities. The limited efficacy and genotoxicity of this drug have hindered its application in clinic. Passive prevention with palivizumab or nirsevimab is restricted to children under two years old and requires high costs of treatment (USD 5000 per children for the winter season)15. Natural products have been a highly valuable and productive source of bioactive molecules for drug development16, 17, 18, 19. Searching for natural products with antiviral activities from medicinal plants is a long-term research interest of our group20, 21, 22, 23.
The plant Cleistocalyx operculatus (Roxb.) Merr. & Perry (Myrtaceae) is an evergreen and flowering tree that is native to southern China, whose buds and leaves are traditionally used as a tonic drink or herbal tea for the treatment of cold, fever, and gastrointestinal disorders24. Phytochemical investigations had revealed that C. operculatus contains an abundance of phenols, terpenoids, and flavonoids25, 26, 27. In previous studies, we found that the petroleum ether-soluble fraction of the ethanol extract of this plant showed significant inhibitory effect against RSV. Guided by in vitro anti-RSV assay, a group of new cinnamoylphloroglucinol-terpene adducts (CPTAs) with unusual hybrid architectures were isolated from the buds and leaves of C. operculatus28, 29, 30. Biogenetically, these novel CPTAs were hypothesized to derive from convergence of ployketide and isoprenoid pathways by assembling terpenoid and cinnamoylphloroglucinol building blocks together. Beyond their unique structural features, some members of CPTAs were demonstrated to exhibit promising anti-RSV activity.
In our continuing research for exploring structurally diverse cinnamoylphloroglucinols with antiviral activities from the titled plant, an in-depth phytochemical investigation was conducted by using BBMN targeted isolation strategy. As a result, eighteen novel CPTAs (1–18, Fig. 1) possessing a variety of carbon skeleton types were discovered and characterized from the petroleum ether-soluble fraction of the ethanol extract of C. operculatus buds. Among the isolates, cleistoperone A (1) is a novel CPTA that possesses an unprecedented macrocyclic carbon framework featuring a unique densely functionalized tricyclo[15.3.1.03,8]heneicosane bridged ring system. Particularly, the highly flexible 6/14/6 tricyclic core of 1 contains two different kinds of stereogenic elements (including five point and three planar chiralities) as well as seven contiguous quaternary carbons. Interestingly, due to the presence of an unusual resonance-assisted hydrogen bonding provided by the enolizable β,β′-triketone system of the 6/14/6 tricyclic core, compound 1 exhibits different bridgehead enol forms in solution (C-1-enol form) and solid-state (C-7-enol form). Cleistoperones B and C (2 and 3) are two new CPTAs with a novel carbon skeleton, in which the (nor)monoterpenoid moiety was fused to the cinnamoyl chain of the cinnamoylphloroglucinol unit via two C–C bonds to form an extra cyclohexene ring. Cleistoperones D–Q (4–17) are a class of dihydropyran ring-containing polycyclic CPTAs belonging to nine new skeleton types, which were formed by cinnamoylphloroglucinol units coupled with various monoterpenoid or sesquiterpenoid moieties. Cleistoperone R (18) is the first example of ether bridged CPTA with an unusual bornane motif. The plausible biosynthetic pathways for 1–18 were also proposed. In addition, all of the isolated CPTAs were evaluated for their in vitro anti-RSV activity. Remarkably, compound 1 showed the most potent inhibitory effect against RSV with an IC50 value of 1.71 ± 0.61 μmol/L (ribavirin, IC50 = 15.00 ± 1.00 μmol/L). Preliminary mechanistic study revealed that 1 suppresses RSV infection possibly by affecting the Akt/mTOR/p70S6K signaling pathway, implying a different antiviral mechanism from ribavirin. Herein, we report the targeted isolation, structure elucidation, biosynthetic pathway hypothesis, and anti-RSV activity of these novel CPTAs.
Figure 1.
Chemical structures of compounds 1–18.
2. Results and discussion
2.1. The BBMN construction and annotation of CPTAs
Our recent investigation demonstrated that phloroglucinol derivatives commonly exhibited a characteristic product ion at m/z 195.065, which could be used as a diagnostic ion for the phloroglucinol building block recognization12. In addition, a comprehensive analysis of the tandem mass spectra of CPTA samples from our in-house compound library revealed that the methoxylated CPTAs generally manifested the characteristic product ions at m/z 193.050 with pronounced intensities, indicating that this product ion could be used as a supplementary indication for the diagnosis of cinnamoylphloroglucinol building blocks (Supporting Information Fig. S1). However, due to their diverse chemical skeletons and post-modifications, the terpenoid building blocks did not show any characteristic fragment information in the LC‒MS2 analysis. Considering that the molecular weight of CPTAs is generally greater than 400 Da (cinnamoylphloroglucinols: ∼297 Da and terpenoids >120 Da), a mass-to-charge ratio screening was used for further accurate recognition of CPTAs. Therefore, a BBMN network of CPTAs from C. operculatus buds was constructed (see the Supporting Information for details).
In the generated BBMN network (Fig. 2), the primary cluster distinctly portrayed the distribution of nodes associated with the two diagnostic ions. Nodes with the product ion at m/z 193.050 mainly located in the top-left region of the network, whereas others were mostly situated in the bottom. Through an in-house library annotation, four known CPTAs were successfully annotated within four subregions of the primary cluster. Notably, a series of unknown nodes, those proximate to the annotated compounds, displayed both significant intensities and divergent mass fragment patterns, suggesting that a number of unusual CPTAs remaining to be explored. Additionally, an in-depth comparison of the MS2 spectrum of node at m/z 583.342 in the unclustered area revealed remarkable differences from the known compounds, indicating the existence of unprecedented CPTAs. In view of the above analysis, an LC‒MS guided isolation for these unknown compounds were conducted, leading to the obtainment of compounds 1–18.
Figure 2.
BBMN for the cinnamoylphloroglucinols from C. operculatus and annotation of the known CPTAs.
2.2. Structure elucidation of compounds 1–18
Cleistoperone A (1) was initially obtained as light yellow oil. The molecular formula C38H46O5 for 1 was determined based on its HRESIMS ion peak at m/z 583.3417 [M + H]+ (calcd for C38H47O5, 583.3418). The UV spectrum of 1 displayed absorption maxima at 200, 234, and 292 nm. The IR spectrum revealed the characteristic absorptions for hydroxy group (3417 cm−1), carbonyl group (1713 and 1650 cm−1), and benzene ring (1619 and 1520 cm−1) functionalities. The 1H and 13C NMR spectra of 1 showed signals corresponding to an enol moiety, four ketone carbonyls, four trisubstituted double bonds, a monosubstituted benzene ring, eight tertiary methyls, four methylenes, four methines, and two quaternary carbons. Comprehensive analysis of the 1D and 2D NMR spectroscopic data allowed the full assignment of all proton and carbon resonances of 1 (Supporting Information Table S1).
Four independent spin coupling systems could be deduced from the 1H–1H COSY spectrum of 1 (Fig. 3A). In the HMBC spectrum of 1, cross-peaks between 1-OH and C-1/C-2/C-6, H3-16 and C-1/C-3, H3-17 and C-3/C-5, H3-18 and C-3/C-5, and H-9 and C-7/C-11/C-15 were observed, suggesting the presence of a 2,4,4-trimethyl-phenylpropanoyl-β-triketone moiety (1a) in 1. This assignment was further verified by comparison of the NMR data assigned to 1a with those of the known compound cleistocaltone A, whose structure was unambiguously elucidated by X-ray crystallography29. In addition, the observation of HMBC correlations between H-2′ and C-4′, H3-9′ and C-2′/C-4′, H3-8′ and C-6′, and H3-10′ and C-6′/C-8′ allowed the establishment of a linear monoterpenoid moiety (1b). Meanwhile, another linear monoterpenoid moiety (1c) could also be deduced from the HMBC correlations between H2-1″ and C-3″, H-4″ and C-2″, H3-9″ and C-1″/C-3″, H-5″ and C-7″/C-10″, H3-10″ and C-7″, and H-8″a and C-7″. The above spectroscopic data suggested that 1 is a novel CPTA containing a 2,4,4-trimethyl-phenylpropanoyl-β-triketone motif and two linear monoterpenoid moieties. The assembly of the three substructures 1a, 1b, and 1c through quadruple C–C bonds (C-2–C-1′, C-8–C-4″, C-9–C-1″, and C-4′–C-8″) was ultimately determined by the key HMBC cross-peaks between H2-1′ and C-1/C-3, along with the 1H–1H COSY correlations of H-8/H-4″, H-9/H-1″a, and H-4′/H-8″a. Thus, the planar structure of 1, possessing an unprecedented tricyclo[15.3.1.03,8]heneicosane-bridged core, was explicitly established (Fig. 3A).
Figure 3.
Structure elucidation of compound 1. (A) Key 1H–1H COSY, HMBC, and NOESY correlations of 1 (the blue solid arrows represent the rotatable direction of the chiral planes). (B) Crystal structure of 1 (thermal ellipsoids are drawn in 50% probability level). (C) Calculated and experimental ECD spectra of 1. (D) Resonance-assisted hydrogen bonding of 1 lead to two different enol forms between solution and solid-state.
As shown in Fig. 3A, compound 1 features a flexible 14-membered macrocyclic scaffold with three isolated stereoclusters (cluster I: C-2, cluster II: C-4′, and cluster III: C-8/C-9/C-4″) and three independent chiral planes (planes a, b, and c), which brought great challenge for its stereostructure assignment. To establish the relative configuration of 1, a NOESY experiment was initially performed. In the NOESY spectrum of 1, cross-peaks between H-1′b and H3-9′, H-3″ and H3-9″, H-4″ and H3-10″, as well as H-5″ and H-8″a were observed, indicating that the Δ2′, Δ2″,3″, and Δ5″ double bonds were E-, Z-, and E-geometries, respectively, and the α,β-unsaturated ketone (C-5″/C-6″/C-7″) adopted a S-trans configuration. Moreover, the observed NOE cross-peaks between H-11/H-15 and H-8, H-8 and H-5″, H-5″ and H-8″a, and H-8″a and H-2′ suggested that these protons were co-facial and arbitrarily assigned as β-oriented. On the contrary, the NOE cross-peaks between H-9 and H-4″ as well as H3-10″ and H-4″ indicated that these protons were α-oriented (Fig. 3A). However, due to the lack of reliable NOE correlations, the relative spatial arrangement of C-2 and chiral plane c in 1 remained unassigned. In order to establish the relative configuration of 1, the theoretical calculations of 13C NMR chemical shifts of the four possible stereoisomers of 1 (1A–1D, Supporting Information Fig. S2A) were performed using the GIAO method with Gaussian 09 software at the mPW1PW91/6-311+G(d,p) level. With a highest correlation coefficient (R2) of 0.9989 and a lowest standard deviation (StDEV) of 2.12 ppm between the predicted and experimental data, the calculated 13C NMR data of 1A were in excellent agreement with the experimental ones. Furthermore, the results of DP4+ analysis31 showed the dominant probability of 100.00% for 1A, permitting the establishment of the relative configuration of 1 as 2S∗,8R∗,9R∗,4′S∗,4″R∗,7RP∗,2′SP∗,5″SP∗ (Fig. S2B–S2D).
Fortunately, after numerous attempts, tiny single-crystals of 1 were finally obtained from an optimized trinary solvent system (MeOH/CH2Cl2/H2O, 7:2:1). Subsequent X-ray diffraction experiment unambiguously confirmed its planar structure and relative configuration (Fig. 3B). To our surprise, the crystallographic structure of 1 (CCDC 2333986) clearly showed that the bridgehead enol was located at C-7/C-6 instead of the C-1/C-6 position established from the NMR spectra of 1, indicating the presence of an intramolecular hydrogen-bonded proton transfer between the solution and solid-state. To investigate this unusual proton transfer phenomenon within hydrogen bonding, the hydrogen-bonded binding energies (BEs) of isomers 1-I (C-1-enol form) and 1-II (C-7-enol form) were calculated using density functional theory (DFT) at the B3LYP-D3(BJ)/ma-TZVPP level. As the results shown in Fig. 3D, two extreme low BE values for 1-I (−15.22 kcal/mol) and 1-II (−14.51 kcal/mol) were obtained, which indicated that a prominent O–H⋯O interaction occurred in the enolizable β,β′-triketone system of 1. This prominent O–H⋯O interaction was also confirmed by the remarkable downfield shift of the enolic proton resonance (18.45 ppm) in the 1H NMR spectrum of 1. Furthermore, in the crystal structure of 1, the bond lengths of C-1–O-1 and C-7–O-4 were measured to be about 1.26 and 1.29 Å, respectively, which are between the standard C–O (1.42 Å) and C O (1.20 Å) bonds. Similarly, bond lengths of C-1–C-6 (1.43 Å) and C-6–C-7 (1.42 Å) are also between the standard C–C (1.54 Å) and C C (1.32 Å) bonds. These unusual bond lengths suggested the occurrence of a strong electronic delocalization in the heteroconjugated HO–C C–C O system of 1 (Fig. 3B). Thus, the synergistic reinforcement of hydrogen bonding and π-delocalization led to the occurrence of a resonance-assisted hydrogen bonding32,33 in 1. As both 1-I and 1-II exhibited extreme low BE values, the exchangeable proton can potentially bond to either of the two adjacent oxygen atoms based on the ambient changes, ultimately forming the different enol forms of 1 in solution and solid-state (Fig. 3D).
The P21/c space group suggested the racemic nature of 1, which was supported by a barely measurable optical rotation value. Subsequently, (±)-1 was separated by chiral HPLC to yield two optically pure enantiomers, (+)-1 and (−)-1, with an equal ratio (Supporting Information Fig. S3). To determine their absolute configurations, quantum chemical ECD calculations for the two possible absolute configurations of 1 were carried out at the PCM/CAM-B3LYP/6-31+G(d) level. The experimental ECD curves of (+)-1 and (−)-1 were well matched with the calculated results of (2S,8R,9R,4′S,4″R,7RP,2′SP,5″SP)-1 and (2R,8S,9S,4′R,4″S,7SP,2′RP,5″RP)-1, respectively (Fig. 3C). Therefore, the absolute configuration of each enantiomer was unambiguously established.
Cleistoperone B (2) possessed a molecular formula C28H32O5, as deduced from its HRESIMS (m/z 449.2330 [M + H]+, calcd for C28H33O5, 449.2323). The UV spectrum of 2 displayed absorption maxima at 200 and 297 nm, indicated the skeletal difference between 2 and 1. Interpretation of 2D NMR spectral data could fully construct the planar structure of 2. In detail, the HMBC correlations between 1-OH and C-1/C-2/C-6, H3-16 and C-1/C-3, H3-17 and C-3/C-5, H3-18 and C-5, H-8 and C-7, and H-11/15 and C-9, combined with the 1H–1H COSY correlations of H-8/H-9 and H-11/H-12/H-13/H-14/H-15, suggested the existence of a phenylpropanoyl-phloroglucinol moiety (2a) in 2. Meanwhile, the observed HMBC cross-peaks between H-2′ and C-4′, H3-9′ and C-2′/C-4′, H-5′b and C-7′, H-8′ and C-6′/C-10′, and H3-10′ and C-6′, along with the 1H–1H COSY correlations of H2-1′/H-2′ and H-4′/H2-5′/H-6′, resulted in the establishment of a linear monoterpenoid unit (2b). Moreover, the 1H–1H COSY correlations of H-8/H-4′ and H-9/H2-1′ indicated that the two fragments 2a and 2b were connected via C-8–C-4′ and C-9–C-1′ bonds to form a cyclohexene ring (Fig. 4).
Figure 4.
Key 1H–1H COSY and HMBC correlations of compounds 2–18.
In the NOESY spectrum of 2, cross-peaks between H-2′ and H3-9′ as well as H-6′ and H-8′ were observed, indicating that the Δ2′ and Δ6′ double bonds were Z- and E-geometries, respectively. In addition, the NOE correlations observed between H-8 and H-11 as well as H-9 and H-5′a established the relative configurations of C-8, C-9, and C-4′ in 2 (Fig. 5). The following X-ray crystallographic analysis (CCDC 2333987) provided the conclusive evidence for above planar structure and relative configuration assignments (Fig. 6). The presence of P space group suggested that 2 was also a racemic mixture. Chiral HPLC resolution of (±)-2 led to the obtainment of two anticipated enantiomers with a ratio of 1:1 (Fig. S3). Finally, the absolute configurations of (+)-2 and (−)-2 were respectively determined as 8S,9S,4′R and 8R,9R,4′S by using a similar ECD calculation method (Supporting Information Fig. S4).
Figure 5.
Key NOESY correlations of compounds 2–17.
Figure 6.
Crystal structures of compounds 2, 3, 14, 16, and 18 (thermal ellipsoids are drawn in 50% probability level).
The HRESIMS data of cleistoperone C (3) exhibited an [M + Na]+ ion peak at m/z 457.1978, suggesting a molecular formula of C27H30O5 (calcd for C27H30O5Na, 457.1985) for 3. Similar to 2, the 1H and 13C NMR spectra of 3 showed characteristic signals for an identical phenylpropanoyl-phloroglucinol moiety (3a). The proton spin coupling system (H-3′/H-4′/H-5′) deduced from the 1H–1H COSY spectrum of 3, combined with the HMBC correlations between H3-8′ and C-1′/C-3′, H-1′b and C-3′, H-5′ and C-7′, H3-9′ and C-5′, and H-7′ and C-6′/C-9′ assembled the remaining proton and carbon resonances into a linear nor-monoterpenoid moiety (3b). Furthermore, the 1H–1H COSY spectrum of 3 revealed the presence of spin coupling systems of H-4′/H-8 and H-1′b/H-9 indicated that substructures 3a and 3b were connected through C-8–C-4′ and C-9–C-1′ bonds (Fig. 4). In the NOESY spectrum, the observed cross-peaks between H-8 and H-11/15, H-9 and H-4′, H-3′ and H3-8′, and H-5′ and H-7′ established the relative configuration of 3 (Fig. 5). Finally, the structure of 3 was unquestionably confirmed by single-crystal X-ray diffraction analysis (Fig. 6, CCDC 2333988). Similarly, compound 3 was also obtained as a racemic mixture according to the P21/c space group. Thus, the racemate of (±)-3 was subsequently separated into two enantiomers by employing a similar chiral HPLC method (Fig. S3). The absolute configurations of (+)-3 and (−)-3 were respectively determined as 8R,9R,4′R and 8S,9S,4′S by comparison of its experimental and calculated ECD curves (Fig. S4).
Cleistoperone D (4) showed a molecular formula of C33H40O4 according to the HRESIMS ion peak at m/z 501.3001 [M + H]+ (calcd for C33H41O4, 501.2999). Unlike 1–3, the UV spectrum of 4 exhibited absorption maxima at 200, 231, 304, and 378 nm, indicating the presence of an extended conjugation system in 4. Comparison of the 1H and 13C NMR spectroscopic data of 4 with those of champanone B34 suggested that 4 possesses an identical 2,4,4-trimethyl-cinnamyl-β-triketone moiety (4a). Based on the 1H–1H COSY correlations of H-1′/H-6′ and H3-12′/H-11′/H3-13′ in 4, as well as the HMBC correlations between H-1′ and C-3′, H-2′a and C-4′/C-6′/C-10′, H-5′ and C-7′, H-6′ and C-4′/C-10′/C-11′, H2-8′ and C-6′, H2-9′ and C-7′, H3-12′/H3-13′ and C-7′, H3-14′ and C-5′/C-9′, and H-15′a and C-4′, the remaining proton and carbon signals were assigned to construct a copane moiety (4b). In addition, the 1H–1H COSY correlation of H2-16/H-15′a indicated the C-16–C-15′ bonded linkage between fragments 4a and 4b. Finally, based on the downfield shift of C-4′ (δC 85.4) and the molecular formula information, a dihydropyran ring was formed by connecting C-3 and C-4′ via an oxygen atom (Fig. 4). Due to the presence of a rigid tricyclo[4.4.0.02,7]decane ring system in 4, the relative configurations of C-1′, C-5′, C-6′, and C-10′ were fixed as 1′S∗,5′R∗,6′R∗,10′S∗ (Supporting Information Fig. S5). Therefore, the obvious NOE cross-peaks between H3-14′ and H-2′a/H2-3′/H-15′b as well as H3-13′ and H-1′/H-6′ established the relative configurations of C-4′, C-7′, and C-1′/C-5′/C-6′/C-10′ in 4 (Fig. 5). Finally, the absolute configuration of 1′S,4′R,5′R,6′S,7′S,10′R for 4 was determined by comparison of its experimental and calculated ECD curves (Fig. S4).
Cleistoperone E (5) was determined to possess the identical molecular formula to that of 4 based on its HRESIMS data. Comparison of the NMR spectra data of 5 with those of 4 suggested that they shared the same 2,4,4-trimethyl-cinnamoyl-β-triketone moiety (5a). Besides, the 1H–1H COSY spectrum of 5 revealed the presence of two additional spin coupling systems (H2-2′/H-3′ and H-9′b/H-10′/H3-14′). The HMBC correlations between H2-2′ and C-1′/C-4′/C-6′, H-9′a and C-1′/C-7′, H-8′a and C-6′, H3-12′ and C-7′, H3-13′ and C-7′/C-12′, H-11′ and C-6′, H-5′ and C-7′, H-10′ and C-5′, H3-14′ and C-1′, and H3-15′ and C-3′/C-5′ allowed the establishment of a cubebane moiety (5b). Similar to 4, based on the 1H–1H COSY correlation of H-16b/H-3′ and the diagnostic downfield shift of C-4′ (δC 88.6), the two substructures 5a and 5b were determined to be linked via C-16–C-3′ and C-3–O–C-4′ bonds (Fig. 4). In the NOESY spectrum of 5, the cross-peaks between H-3′ and H3-15′/H-6′ as well as H-6′ and H3-13′ assigned those protons on the same side of the molecule. In contrast, the NOE correlations between H-5′ and H-7′/H-9′b as well as H3-14 and H-9′b/H3-17′ located those protons on the other orientation of the molecule (Fig. 5). Finally, the absolute configuration of 5 was determined to be 1′R,3′S,4′R,5′R,6′R,7′S,10′R by comparing its experimental and calculated ECD data (Fig. S4).
Cleistoperone F (6) shared the same molecular formula of C33H40O4 as 5, as inferred from its HRESIMS. Comparison of the 1H and 13C NMR spectral data of 6 with those of 5 revealed that 6 contained the identical 2,4,4-trimethyl-cinnamoyl-β-triketone moiety (6a) to that of 5. Interpretation of the 1H–1H COSY spectrum of 6 led to the establishment of a large spin coupling system of H2-9′/H-8′a/H-7′/H-6′/H-5′/H-4′/H2-3′(H3-15′)/H-2′a/H-1′/H-5′. In the HMBC spectrum, the correlations between H-1′ and C-14′, H-5′ and C-10′/C-11′, H-8′a and C-10′, H3-12′ and C-6′/C-7′/C-13′, and H3-13′ and C-6′/C-7′ assembled a 5/7/3 tricyclic aromadendrane moiety (6b). In addition, the 1H–1H COSY correlation of H-16a/H-14′b indicated that substructures 6a and 6b were connected via C-16–C-14′ bond (Fig. 4). Accordingly, the formation of oxygen bridge between C-3 and C-10′ was determined by the molecular formula information and the obvious downfield shift of C-10′ (δC 85.7). In the NOESY spectrum of 6, the key cross-peaks between H-1′ and H3-15′/H-6′ as well as H3-13′ and H-6′/H-7′ suggested that these protons were co-facial and assigned as β-oriented. Meanwhile, the observed NOE cross-peaks between H-5′ and H3-12′/H-14′a indicated that these protons were defined as α-oriented (Fig. 5). The absolute configuration of 6 was finally identified as 1′R,4′R,5′S,6′S,7′R,10′S by employing the quantum chemical ECD calculation (Fig. S4).
Cleistoperone G (7) was deduced to possess a molecular formula C33H36O4 according to the protonated adduct ion peak at m/z 497.2684 in its HRESIMS (calcd for C33H37O4, 497.2686). Similar to 4–6, the 1H and 13C NMR spectra of 7 exhibited typical signals due to a 2,4,4-trimethyl-cinnamoyl-β-triketone moiety (7a). Besides, the two spin coupling systems [H-4′/H-5′ and H-8′/H-9′a/H-10′/H-11′/H3-12′ (H3-13′)] interpreted from the 1H–1H COSY spectrum of 7, together with the observed HMBC correlations between H-2′ and C-4′/C-10′/C-14′, H-4′ and C-6′/C-14′, H-5′ and C-1′/C-3′/C-7′, H-8′a and C-6′, H-9′a and C-1′/C-7′, and H-15′a and C-6′/C-8′ led to the establishment a calamenene moiety (7b) in 7. Furthermore, the 1H–1H COSY correlation of H-16a/H-15′a indicated that the two fragments 7a and 7b were linked via C-16–C-15′ bond. Similarly, based on the obvious downfield shift of C-7′ (δC 80.4) and the molecular formula information, the remaining unassigned oxygen atom was deduced to bridge C-3 and C-7′ to form a dihydropyran ring (Fig. 4). In the NOESY spectrum of 7, cross-peaks between H-9′b and H-15′a/H3-13′, H-5′ and H-15′b, and H-10′ and H3-12′ were observed, allowing the establishment of the relative configurations of C-7′ and C-10′ (Fig. 5). Finally, based on the ECD calculation results, the absolute configuration of 7 was established to be 7′S,10′S (Fig. S4).
Cleistoperone H (8) was determined to have the molecular formula C33H40O4 on the basis of its HRESIMS data. Similar to 4–7, the 1H and 13C NMR spectral data of 8 showed characteristic proton and carbon resonances for a 2,4,4-trimethyl-cinnamoyl-β-triketone moiety (8a). For the sesquiterpenoid moiety, three spin coupling systems of H-1′/H-2′/H-3′a, H-5′/H-6′a/H-7′b, and H-9′/H-10′a could be deduced from the 1H–1H COSY spectrum of 8. Furthermore, in the HMBC spectrum of 8, the observed correlations between H3-12′ and C-3′/C-5′, H-3′a and C-5′, H-6′a and C-8′, H-7′a and C-9′, H3-13′ and C-7′/C-9′, H3-14′ and C-1′/C-10′/C-15′, H3-15′ and C-1′/C-10′, and H-10′b and C-1′ allowed the establishment of a macrocyclic α-humulene moiety (8b). Finally, the 1H–1H COSY correlation of H-16a/H-5′ and the obvious downfield shift of C-4′ (δC 82.5) led to the construction of a dihydropyran ring between substructures 8a and 8b (Fig. 4).
In the NOESY spectrum of 8, cross-peaks between H-1′ and H-3′b, H-10′a and H3-13′, and H-7′b and H-9′ were observed, indicating that the double bonds at Δ1′ and Δ8′,9′ were both assigned as E-geometry. In addition, crucial NOE cross-peaks between H3-12′ and H-16b/H-6′b were observed, suggesting these protons as being on the same side of the molecule. On the contrary, the NOE cross-peak between H-5′ and H-3′b suggested that the two protons were located on the other side of the molecule (Fig. 5). The optical rotation value of 8 was close to zero, suggesting that 8 was also a racemic mixture. The racemate of (±)-8 was further resolved by chiral HPLC to afford a pair of enantiomers (Fig. S3). Similarly, the absolute configurations of (+)-8 and (−)-8 were then assigned as 4′S,5′R and 4′R,5′S, respectively, by comparing their experimental and calculated ECD curves (Fig. S4).
Cleistoperone I (9) showed the same molecular formula C33H40O4 as 8 by its HRESIMS data. Similar to 8, the 1H and 13C NMR spectra of 9 showed characteristic signals due to an α-humulene moiety (9b). Different from 8, the spin coupling systems (H-8/H-9 and H-11/H-12/H-13/H-14/H-15) obtained from the 1H–1H COSY spectrum of 9 in conjunction with the HMBC correlations between 1-OH and C-1/C-2/C-6, H-16a and C-2, H3-17 and C-3/C-5, H3-18 and C-5, H-8 and C-10, and H-9 and C-7/C-11/C-15 revealed the presence of a 2,4-dimethyl-cinnamoylphloroglucinol moiety (9a) in 9 instead of the 2,4,4-trimethyl-cinnamoyl-β-triketone moiety in 8. Furthermore, based on the 1H–1H COSY correlation of H-16a/H-5′ and the diagnostic downfield shift of C-4′ (δC 82.5), the coupling pattern between fragments 9a and 9b via C-16–C-5′ and C-3–O–C-4′ bonds were also confirmed to be identical to that of 8 (Fig. 4). Detailed analysis of the NOESY spectrum revealed that 9 possessed the same relative configurations at C-4′ and C-5′ positions as those in 8 (Fig. 5). Due to its barely measurable optical rotation, this suggested that 9 was also isolated as a racemic mixture. Further chiral HPLC separation of 9 afforded two anticipated enantiomers (Fig. S3). Finally, the absolute configurations of (−)-9 and (+)-9 were determined as 4′R,5′S and 4′S,5′R, respectively, by comparing their experimental and calculated ECD spectra (Fig. S4).
The molecular formula of cleistoperone J (10) was established as C33H40O4 on the basis of its HRESIMS data. Similar to 4–8, the 1H and 13C NMR spectra of 10 also revealed the presence of a 2,4,4-trimethyl-cinnamoyl-β-triketone moiety (10a). The 1H–1H COSY correlations of H-1′/H-9′/H2-10′, H-2′a/H-3′a, and H-5′/H-6′a/H-7′a in conjunction with the HMBC cross-peaks between H-3′b and C-1′, H2-2′ and C-4′, H3-14′ and C-3′/C-5′, H-6′a and C-4′, H-7′a and C-9′/C-15′, H-9′ and C-15′, H3-12′ and C-1′/C-10′, and H3-13′ and C-1′/C-10′/C-12′ established a β-caryophyllene moiety (10b) in 10. In addition, the key HMBC correlations between H-16′a and C-4′/C-6′ indicated that the two fragments 10a and 10b were connected via C-16–C-5′ bond. Based on the obvious downfield shift of C-4′ (δC 84.3) as well as the molecular formula information of 10, an oxygen bridge was further formed between C-3 and C-4′ (Fig. 4). In the NOESY spectrum of 10, the observed NOE cross-peaks between H3-14′ and H-16b/H-6′a/H-7′b, H-9′ and H-7′b/H3-13′, and H3-12′ and H-1′ indicated that H3-14′, H-9′, and H3-13′ were co-facial, while H-1′, H-5′, and H3-12′ were on the other side of the molecule (Fig. 5). Finally, based on a good agreement between the calculated ECD curve of 1′R,4′R,5′S,9′S-10 and the experimental one, the absolute configuration of 10 was established (Fig. S4).
Cleistoperone K (11) was found to have the same molecular formula as 10 based on its HRESIMS data. Further detailed analysis of the NMR data revealed that 11 possessed the identical planar structure to 10. Different from 10, the NOESY spectrum of 11 showed correlations between H-1′ and H-5′/H-12′, suggesting that these protons were co-facial and identified as β-orientated. On the contrary, the NOE cross-peaks between H3-13′ and H-9′ as well as H3-14′ and H-16b/H-6′b indicated that these protons were α-orientated (Fig. 5). The above data suggested that 11 possessed the opposite configurations to 10 at the C-4′ and C-5′ positions. Similarly, the calculated ECD curve for 1′R,4′S,5′R,9′S-11 resembled the experimental one. Thus, the absolute configuration of 11 was determined (Fig. S4).
The molecular formular of cleistoperone L (12) was assigned as C33H40O4 on the basis of its HRESIMS data. Compound 12 was determined to possess the identical β-caryophyllene moiety (12b) to that of 10 and 11 by comparison of their 1H and 13C NMR spectral data. Besides, the 1H–1H COSY correlations of H-8/H-9 and H-11/H-12/H-13/H-14/H-15, together with the HMBC cross-peaks between 1-OH and C-1/C-2/C-6, H-16a and C-1/C-3, H3-17 and C-3/C-5, H3-18 and C-5, H-8 and C-10, and H-9 and C-7/C-11/C-15 established a 2,4-dimethyl-cinnamoylphloroglucinol moiety (12a) in 12. The connection of fragments 12a and 12b through C-16–C-5′ and C-3–O–C-4′ bonds was confirmed to be identical to that of 11 (Fig. 4). In the NOESY spectrum of 12, correlations between H3-14′ and H-16b/H-6′a as well as H-9′ and H-6′a/H3-13′ were observed, suggesting that these protons were co-facial and identified as β-orientated. Meanwhile, the NOE cross-peak between H-1′ and H3-12′ indicated the α-orientation of these protons (Fig. 5). Finally, the absolute configuration of 12 was determined as 1′R,4′R,5′S,9′S by comparing its experimental and calculated ECD spectra (Fig. S4).
Cleistoperone M (13) exhibited an identical molecular formula to 12 from its HRESIMS data. The similarity between the 1H and 13C NMR data of 13 and those of 12 indicated that the two compounds had the identical planar structure, which was corroborated via detailed analysis of the 2D NMR data of 13. In the NOESY spectrum of 13, the cross-peaks between H-1′ and H-5′/H3-12′ indicated that these protons were co-facial and were assigned as β-oriented. In contrast, the observed NOE cross-peaks between H3-14′ and H-16b/H-6′b as well as H3-13′ and H-9′ suggested that H-6′b, H-9′, H3-13′, H3-14′, and H-16b were presented in the α-orientation (Fig. 5). Thus, compound 13 was identified as a stereoisomer of 12, with opposite configurations to 12 at the C-4′ and C-5′ positions. Finally, the absolute configuration of 13 were determined to be 1′R,4′S,5′R,9′S by comparison of the experimental and calculated ECD spectra (Fig. S4).
Cleistoperone N (14) showed the same molecular formula C33H40O4 as 12 and 13 based on its HRESIMS data. Similar to 12 and 13, the 1D and 2D NMR spectra of 14 showed characteristic signals due to a 2,4-dimethyl-cinnamoylphloroglucinol moiety (14a) and a β-caryophyllene moiety (14b). Different from 12 and 13, the 1H–1H COSY correlation of H-17a/H-5′ indicated that the two substructures 14a and 14b were linked via C-17–C-5′ bond in 14, instead of the C-16–C-5′ linkage in 12 and 13 (Fig. 4). Based on the characteristic downfiled shift of C-4′ (δC 82.1) and the upfiled shift of C-3 (δC 159.5), the remaining oxygen atom was assigned to bridge C-3 and C-4′ to form a dihydropyran ring. Finally, the gross structure of 14 was unambiguously confirmed by X-ray crystallography (CCDC 2333989). With a Flack parameter of 0.05(19), the absolute configuration of 14 was defined as 1′R,4′S,5′R,9′S (Fig. 6).
Cleistoperone O (15) showed an identical molecular formula to 14 from its HRESIMS data. The 1H and 13C NMR spectroscopic data of 15 were found to be very similar to those of 14, except for minor differences of signals assigned to protons and carbons located in the connecting dihydropyran ring. Further analysis of the 1H–1H COSY, HSQC, and HMBC data of 15 indicated that the two compounds possessed identical planar structures (Fig. 4). Different from 14, the NOESY spectrum of 15 showed cross-peaks between H-1′ and H-5′/H3-13′, indicating that these protons were co-facial and assigned as α-oriented. Conversely, the cross-peaks between H3-12′ and H-9′ as well as H-6′a and H-9′/H3-14′ allowed the assignment of β-orientation for all of these protons. Thus, the above data suggested that 15 was a C-4′ and C-5′ isomer of 14 (Fig. 5). Similarly, the absolute configuration of 15 was determined as 1′R,4′R,5′S,9′S by comparing its experimental and calculated ECD curves (Fig. S4).
The molecular formula of cleistoperone P (16) was determined to be C28H32O4 on the basis of its HRESIMS data (m/z 433.2371 [M + H]+, calcd for C28H33O4, 433.2373). Similar to 4, the 1H and 13C NMR spectra of 16 showed characteristic signals due to a 2,4,4-trimethyl-cinnamoyl-β-triketone moiety (16a). Unlike 4, detailed analysis of the 1H–1H COSY, HSQC, and HMBC spectra of 16 revealed that the remaining NMR signals could be attributed to a α-pinene moiety (16b). The connection between fragments 16a and 16b through C-16–C-8′ was deduced by the 1H–1H COSY correlation of H2-16/H-8′b. Moreover, the typical downfield shift of C-2′ (δC 85.5) and the molecular formula information allowed the construction of a dihydropyran ring between 16a and 16b via C-3–O–C-2′ bond (Fig. 4). Finally, the intact structure of 16 were unambiguously confirmed based on an X-ray crystallographic experiment (Fig. 6, CCDC 2333990). Although a specific optical rotation value was observed in 16, the presence of a symmetric P21/n space group in the X-ray structure suggested that 16 was obtained as a partially racemic mixture. After chiral HPLC separation, a pair of enantiomers, (+)-16 and (−)-16, with a ratio of 87:13, were obtained (Fig. S3). Using a similar ECD calculation method, the absolute configurations of (−)-16 and (+)-16 were established as 1′S,2′S,5′R and 1′R,2′R,5′S, respectively (Fig. S4).
The molecular formula of cleistoperone Q (17) was determined to be identical to that of 16 by its HRESIMS data. Similar to 16, the 1H and 13C NMR spectra of 17 showed feature signals due to a 2,4,4-trimethyl-cinnamoyl-β-triketone unit (17a) and an α-pinene moiety (17b). Different from 16, the 1H–1H COSY correlation of H-16a/H-3′, the HMBC cross-peak between H-16a and C-2′, together with the downfield chemical shift of C-2′ (δC 85.5) revealed that in 17 the two substructures 17a and 17b were connected via C-16–C-3′ and C-3–O–C-2′ bonds instead of the C-16–C-8′ and C-3–O–C-2′ bonds in 16 (Fig. 4). In the NOESY spectrum of 17, cross-peaks between H-3′ and H3-8′/H3-10′, H3-8′ and H3-10′, and H3-9′ and H-7′a were observed, allowing the establishment of the relative configuration of 17 (Fig. 5). Similar to 16, compound 17 was also a partially racemic mixture. By using a chiral HPLC separation, a pair of optically pure enantiomers, (−)-17 and (+)-17, in a ratio of 80:20 were obtained (Fig. S3). The theoretical ECD curves generated for 1′R,2′S,3′R,5′S-17 and 1′S,2′R,3′S,5′R-17 were in good agreement with experimental ones for (−)-17 and (+)-17, respectively, which led to the determination of the absolute structures of (−)-17 and (+)-17 (Fig. S4).
Cleistoperone R (18) showed a molecular formula of C28H34O4 by HRESIMS at m/z 435.2357 [M + H]+ (calcd for C28H35O4, 435.2350). Similar to 16 and 17, the 1H and 13C NMR spectra of 18 showed characteristic proton and carbon resonances corresponding to a 2,4,4-trimethyl-cinnamoyl-β-triketone moiety (18a). The 1H–1H COSY spectrum of 18 revealed the presence of two spin coupling systems (H-3′/H-4′a/H-5′ and H-6′a/H-7′b). In the HMBC spectrum, the correlations between H-3′ and C-5′, H-4′b and C-2′, H-5′ and C-6′, H-6′a and C-2′, H3-8′ and C-5′/C-6′, H3-9′ and C-1′/C-3′, and H3-10′ and C-1′/C-3′/C-9′ allowed the establishment of a bornane moiety (18b). Different from 16 and 17, the key HMBC cross-peak between H-5′ and C-3 as well as the downfield shift of C-5′ (δC 87.5) indicated that the two fragments 18a and 18b were linked via C-3–O–C-5′ bonds instead of a dihydropyran ring in 16 and 17 (Fig. 4). The proposed structure of 18 were further confirmed by the following X-ray crystallographic analysis (Fig. 6, CCDC 2333991). The centrosymmetric P space group implied that 18 was obtained as a racemic mixture. Subsequently, the racemic mixture was separated by chiral HPLC to yield two pure enantiomers in a ratio of 1:1 (Fig. S3). Using a similar ECD calculation method, the absolute configurations of (+)-18 and (−)-18 were determined as 1′S,3′R,5′S and 1′R,3′S,5′R, respectively (Fig. S4).
2.3. Plausible biosynthetic pathways of compounds 1–18
Compounds 1–18 represent a collection of biogenetically-related novel CPTAs, which characterized by a polymethylated cinnamoylphloroglucinol building block fused to diverse terpene building blocks in different coupling patterns. The hypothetical biosynthetic pathways for these novel CPTAs were illustrated in Scheme 1, Supporting Information Schemes S1 and S2. Initially, the cinnamoyl-β-triketone could be derived from a characteristic polyketide (PKS) pathway involving the condensation of one cinnamoyl-CoA and three malonyl-CoA units. Enolization or aromatization of the cinnamoyl-β-triketone followed by methylation could lead to the generation of two phloroglucinol building blocks, champanone B (19) and 2′,4′-dihydroxy-6′-methoxy-3′,5′-dimethylchalcone (20), respectively28,35. Addition of geranyl pyrophosphate to phloroglucinol building block 19 could give birth to racemic intermediate i. The allylic hydroxylation product of i was also isolated as a natural product in our previous study29. As a dienophile, intermediate i could further couple with linear monoterpene building block cosmene via Diels–Alder cycloaddition36 to form intermediate ii. Subsequently, the allylic hydroxylation of ii could yield intermediate iii. Protonation of hydroxy group and dehydration of iii provided an allylic cation intermediate iv. Then, a crucial intramolecular electrophilic cyclization of iv followed by quenching with H2O would construct the unprecedented tricyclo[15.3.1.03,8]heneicosane bridged ring scaffold of intermediate v. Finally, oxidation of the newly formed hydroxy group of v could afford compound 1. On the other hand, phloroglucinol building block 20 could also incorporate different linear monoterpene building blocks (cosmene and ocimene) through hetero-Diels–Alder reaction to generate intermediates vi and vii, respectively. Then, oxidation at gem-methyl group of vi could give compound 2, while oxidative cleavage37 of the terminal double bond in vii would generate compound 3. Meanwhile, oxidative activation38 of building blocks 19 or 20 afforded highly reactive β-triketone intermediate Si or phloroglucinol intermediates Sii and Siii (Schemes S1 and S2). As enophiles, intermediates Si, Sii or Siii could further cyclize with terpene building blocks copane, cubebene, aromadendrene, calacorene, humalene, β-caryophyllene, (±)-α-pinene, or (±)-β-pinene via a hetero-Diels–Alder cycloaddition39,40 to generate the corresponding compounds 4–17. Interestingly, chiral HPLC analysis revealed that both compounds 16 and 17 existed as scalemic mixtures, which may be attributed to the partially racemic nature of pinene in plant41. Finally, the p-menthene might undergo intramolecular cyclization to generate bicyclic bornyl cation, which could be trapped by nucleophilic hydroxy group of 19 to produce compound 18.
Scheme 1.
Proposed biosynthetic pathway of compounds 1–3.
2.4. Antiviral activity of compounds 1–18
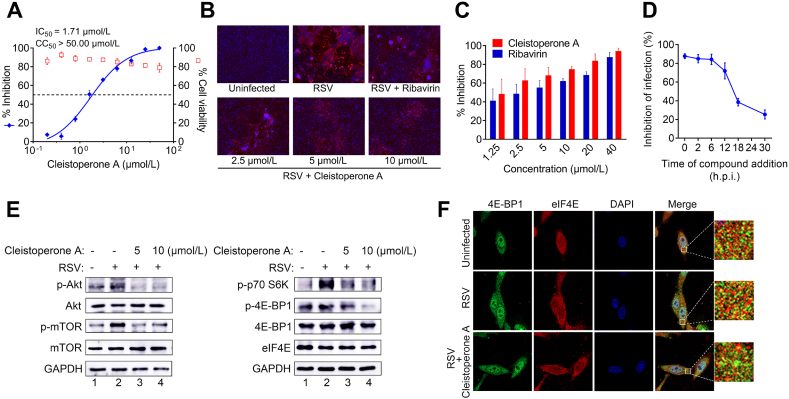
In this study, all of the isolated CPTAs were evaluated for their in vitro antivirus activity against RSV by using a cytopathic effect (CPE) reduction assay. As shown in Table 1, compounds with the presence of cinnamoyl-β-triketone motifs, such as compounds 1, 4–8, 10, 11, and 18, exhibited obvious anti-RSV activities with IC50 values in the range of 1.71–31.25 μmol/L, suggesting that the cinnamoyl-β-triketone motif is essential for the antivirus activity of CPTAs. Exceptionally, compounds 16 and 17 with a cinnamoyl-β-triketone motif did not show anti-RSV activity (IC50 > 50 μmol/L) at the tested concentrations. This could probably be explained by their significant differences in the terpene motifs. Among them, compound 1 showed the most potent anti-RSV activity with IC50 value of 1.71 ± 0.61 μmol/L (Fig. 7A). Furthermore, the immunofluorescence analysis demonstrated that 1 significantly decreased the expression level of RSV fusion protein in a dose-dependent manner (Fig. 7B and C). Notably, no apparent cytotoxicity was observed in HEp-2 cells that were treated with 1 at the concentrations capable of reducing RSV infection.
Table 1.
In vitro anti-RSV activity of compounds 1–18.
| Compound | IC50 ± SD (μmol/L)a | CC50 ± SD (μmol/L)b |
|---|---|---|
| 1 | 1.71 ± 0.61 | >50 |
| 2 | 50 | >50 |
| 3 | 50 | >50 |
| 4 | 6.32 ± 0.69 | 26.61 ± 5.02 |
| 5 | 31.25 ± 8.75 | >50 |
| 6 | 23.75 ± 1.25 | >50 |
| 7 | 12.57 ± 0.92 | >50 |
| 8 | 13.14 ± 1.23 | 47.28 ± 3.23 |
| 9 | 49.55 ± 3.16 | >50 |
| 10 | 25.75 ± 3.75 | >50 |
| 11 | 24.67 ± 1.74 | >50 |
| 12 | 50 | >50 |
| 13 | 50 | >50 |
| 14 | 50 | >50 |
| 15 | 50 | >50 |
| 16 | >50 | >50 |
| 17 | >50 | >50 |
| 18 | 3.13 ± 0.69 | 44.47 ± 2.49 |
| Ribavirinc | 15.00 ± 1.00 | >50 |
IC50: 50% inhibition concentration.
CC50: 50% cytotoxicity concentration.
Positive control: Ribavirin.
Figure 7.
The antiviral activity of 1 against RSV. (A) The inhibitory effect of different concentrations of 1 on RSV infection as determined by CPE reduction assay. (B) Representative images of immunofluorescence analysis from the confocal microscopy: 1 significantly inhibited the expression of virus proteins in a dose-dependent manner. (C) Densitometric analysis for the confocal assay. (D) Inhibition of RSV infection by the treatment of 1 to the HEp-2 cells at different time points after viral infection. (E) Western blot assay. (F) Immunofluorescence analysis of the colocalization between 4E-BP1 and eIF4E using confocal microscopy.
Subsequently, a time-of-addition assay was performed to investigate which stage in the viral life cycle was inhibited by compound 1. As described in Fig. 7D, the virus yield was potently suppressed when 1 was added to cells from 0 to 12 h postinfection (h.p.i.). The inhibitory effect of 1 on RSV infection was rapidly attenuated when the infected cells were treated with 1 after 12 h.p.i. These results suggested that 1 may exert anti-RSV effect in the middle to late stages during the virus replication cycle after the viral entry into host cells. Furthermore, we observed that Akt/mTOR/p70S6K, a signaling pathway involved in the translation of viral RNA, was activated in RSV-infected HEp-2 cells (Fig. 7E). The expression levels of phosphorylated-Akt, -mTOR, and -p70S6K were up-regulated in RSV-infected cells and decreased in the cells that were treated with 1 at 5 or 10 μmol/L. By contrast, the cellular expression of non-phosphorylated Akt, mTOR, and p70S6K in uninfected cells was comparable to that of RSV-infected cells either treated with or without 1. In HEp-2 cells treated with 10 μmol/L of 1, the p-mTOR-mediated phosphorylation of 4E-PB1 was down-regulated, however, the colocalization between eIF4E and 4E-PB1 in these cells was maintained at a comparable level as compared with untreated cells (Fig. 7F). Collectively, these data suggested that 1 inhibits the RSV infection presumably through suppressing the Akt/mTOR/p70S6K signaling pathway. Therefore, 1 represents a privileged scaffold with unusual antiviral mechanism for further development of new anti-RSV agents.
3. Conclusions
In summary, by employing the BBMN-based prioritization approach, eighteen novel CPTAs with diverse skeleton types were isolated and characterized from the buds of medicinal plant C. operculatus. Structurally, these CPTAs were involved in four different kinds of coupling patterns between cinnamoylphloroglucinol and terpene building blocks. Cleistoperone A (1) containing a densely functionalized tricyclo[15.3.1.03,8]heneicosane bridge ring skeleton, represents the first macrocyclic CPTA composed of a cinnamoylphloroglucinol motif and two linear monoterpene moieties. Cleistoperones B and C (2 and 3) are two new skeletal CPTAs with a unique coupling pattern between the (nor)monoterpene moiety and the cinnamoyl chain of the cinnamoylphloroglucinol unit. Cleistoperones D–Q (4–17) represent a class of novel dihydrofuran ring-connected polycyclic CPTAs and 4 possesses an unprecedented cage-like 6/6/6/4/6-fused pentacyclic backbone. Cleistoperone R (18) is the first example of ether bridged CPTA featuring an unusual bornane motif. The discovery of 1–18 not only extremely enriches the chemical diversity of natural phloroglucinols, but also further demonstrates the strong potential of the BBMN approach for rapid and efficient discovery of novel natural products. More encouragingly, the unprecedented chemical structure confers compound 1 with potent anti-RSV activity. The preliminary mechanism of 1 involved inhibiting virus replication by affecting the Akt/mTOR/p70S6K signaling pathway, suggesting that 1 represents a new class of anti-RSV scaffold with a different mode of action from ribavirin. These results shed new lights on the searching for new lead compounds against RSV infection from natural sources.
4. Experimental
4.1. General experimental procedures
The melting points were obtained on an X-5 micromelting apparatus (Fukai Instrument, Beijing, China) without correction. Optical rotation values were measured on a JASCO P-1020 polarimeter (JASCO, Tokyo, Japan) at room temperature. UV and ECD spectra were determined on a Chirascan-plus circular dichroism spectrometer (Applied Photophysics, Leatherhead, Surrey, UK). IR spectra were recorded on a JASCO FT/IR-480 plus Fourier Transform infrared spectrometer (JASCO, Tokyo, Japan) using KBr pellets. HRESIMS data were collected on an Agilent 6210 ESI/TOF mass spectrometer (Agilent, Pala Alto, CA, USA). NMR spectral data were collected on Bruker AV-500 and AV-400 spectrometers (Bruker, Karlsruhe, Germany) using TMS as internal standard. TLC analyses were carried out by using pre-coated silica gel GF254 plates (Yantai Chemical Industry Research Institute, Yantai, China). Column chromatographies were performed on Silica gel (200–300 mesh, Qingdao Marine Chemical Plant, Qingdao, China), ODS gel (Merck, Darmstadt, Germany), MCI gel (CHP20P, 75–150 μm, Mitsubishi Chemical Industries Ltd., Kyoto, Japan), and Sephadex LH-20 (Pharmacia Biotech AB, Uppsala, Sweden). The analytical HPLC analyses were performed on an Agilent 1260 instrument equipped with multiple wavelength diode array detector (DAD), accompanied by a Cosmosil 5C18-MS-II column (4.6 mm × 250 mm, i.d. 5.0 μm, Nacalai Tesque Inc., Kyoto, Japan) or a Phenomenex Luna PFP column (4.6 mm × 250 mm, i.d. 5.0 μm, Phenomenex Inc., Los Angeles, CA, USA). Preparative HPLC preparations were operated on an Agilent 1260 system equipped with 1260 MWD detector, accompanied by a Cosmosil 5C18-MS-II column (20 mm × 250 mm, i.d. 5.0 μm, Nacalai Tesque Inc, Tokyo, Japan) or a Phenomenex Luna PFP column (21.2 mm × 250 mm, i.d. 5.0 μm, Phenomenex Inc., Los Angeles, CA, USA). All solvents used in column chromatography and HPLC were of analytical grade (Tianjin Damao Chemical Plant, Tianjin, China) or chromatographic grade (Merck, Darmstadt, Germany), respectively.
4.2. Plant material
The buds of Cleistocalyx operculatus (Roxb.) Merr. & Perry were collected in May 2021 from Guilin city of Guangxi Zhuang autonomous region (longitude 110°24′ E and latitude 25°62′ N), China. The plant authentication was conducted by Prof. Guangxiong Zhou (College of Pharmacy, Jinan University). A voucher specimen (accession no. CO-20210526) was deposited in the Center for Bioactive Natural Molecules and Innovative Drugs Research, Jinan University.
4.3. Extraction and isolation
The air-dried buds of C. operculatus (20.0 kg) were powdered and extracted with 95% (v/v) ethanol for three times under room temperature. The solution was vacuum-concentrated to give a dark EtOH crude extract (3.1 kg). The crude extract was suspended in water and partitioned successively with petroleum ether, CH2Cl2, and n-butanol. The petroleum ether-soluble fraction was selected and analyzed by UHPLC‒MS2 (positive-ion mode) prior to purification to build BBMN for CPTAs.
The petroleum ether-soluble fraction (806.0 g) was subjected to a silica gel column (200–300 mesh), eluted with petroleum ether/EtOAc (100:0 to 0:100, v/v) to afford fractions A–I. The fraction B (76.1 g) was then chromatographed on an MCI gel column eluted with MeOH/H2O gradient (60:40 to 100:0, v/v), to afford subfractions Ba–Bf. The subfractions Bb (17.3 g) and Bd (15.1 g) were further purified by silica gel column (300–400 mesh), Sephadex LH-20 column (MeOH), and preparative HPLC (MeOH/H2O or MeCN/H2O) to produce compounds 1 (9.1 mg), 4 (5.8 mg), 5 (10.3 mg), and 6 (7.3 mg), respectively. The fraction D (94.4 g) was subjected to a silica gel column (300−400 mesh), eluting with a gradient mixture of cyclohexane/MeOH (100:0 to 100:30, v/v) to give subfractions Da–Dh. The subfractions De (11.8 g) and Dd (18.5 g) were separated by Sephadex LH-20 column, ODS column, and then further purified by preparative HPLC to give compounds 7 (9.4 mg), 8 (8.4 mg), 9 (6.2 mg), 10 (30.2 mg), 11 (20.0 mg), 12 (19.8 mg), 13 (32.7 mg), 14 (6.8 mg), and 15 (5.5 mg), respectively. The fraction E (105.3 g) was subjected to an MCI gel column employing MeCN/H2O (40:60 to 100:0, v/v) as mobile phase to provide subfractions Ea–Ef. The subfraction Ec (9.0 g) was fractionated by Sephadex LH-20 column (MeOH) and further purified by preparative HPLC to afford compounds 2 (6.3 mg) and 3 (7.1 mg), respectively. The subfraction Ee (18 g) was separated via ODS gel column eluted with MeOH/H2O (50:50 to 100:0, v/v), then, further separated by a Sephadex LH-20 column (CH2Cl2/MeOH, 20:80, v/v) and preparative HPLC to obtain compounds 16 (18.0 mg), 17 (13.9 mg), and 18 (11.5 mg), respectively. The chiral separation details and results are presented in the Supporting Information.
4.4. Physico-chemical constants of 1–18
Cleistoperone A (1): light yellow block-shaped crystals (MeOH/CH2Cl2/H2O, 7:2:1); ±0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.81), 234 (3.42), 292 (2.98) nm (Supporting Information Fig. S6); IR 3417, 2975, 2930, 1713, 1650, 1619, 1574, 1520, 1450, 1030, 982, 756, 697, 504 cm−1 (Supporting Information Fig. S7); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S8–S13 and Table S1; HRESIMS m/z 583.3417 [M + H]+ (calcd for C38H47O5, 583.3418) (Supporting Information Fig. S14).
(+)-1: +84.6 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 206 (−4.22), 234 (+16.02), 252 (+20.89), 280 (−6.92), 305 (−9.81) nm.
(−)-1: −83.4 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 205 (+3.15), 234 (−16.93), 251 (−20.47), 282 (+6.21), 304 (+9.31) nm.
Cleistoperone B (2): colorless block-shaped crystals (MeOH/CH2Cl2, 8:2); mp 196–197 °C; ±0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.56), 297 (3.31) nm (Supporting Information Fig. S15); IR 3473, 2927, 1656, 1625, 1509, 1466, 1447, 1201, 1056, 892, 756, 620 cm−1 (Supporting Information Fig. S16); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S17–S22 and Table S2; HRESIMS m/z 449.2330 [M + H]+ (calcd for C28H33O5, 449.2323) (Supporting Information Fig. S23).
(+)-2: +36.3 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 218 (−26.37), 253 (−6.56), 303 (+10.43) nm.
(−)-2: −37.4 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 218 (+25.71), 252 (+6.05), 300 (−11.93) nm.
Cleistoperone C (3): colorless plate-shaped crystals (MeOH/CH2Cl2, 9:1); mp 231–232 °C; ±0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.52), 302 (3.32) nm (Supporting Information Fig. S24); IR 3476, 2941, 1656, 1622, 1514, 1447, 1203, 1144, 1062, 887, 756, 623 cm−1 (Supporting Information Fig. S25); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S26–S31 and Table S2; HRESIMS m/z 457.1978 [M + Na]+ (calcd for C27H30O5Na, 457.1985) (Supporting Information Fig. S32).
(+)-3: +41.1 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 212 (+18.10), 249 (+10.99), 300 (−3.62) nm.
(−)-3: −42.0 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 212 (−19.63), 249 (−12.41), 302 (+3.24) nm.
Cleistoperone D (4): yellow oil; −87.9 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.51), 231 (3.06), 304 (3.00), 378 (3.37) nm (Supporting Information Fig. S33); IR 3456, 2936, 2867, 1644, 1631, 1571, 1407, 1195, 1172, 1113, 751, 700 cm−1 (Supporting Information Fig. S34); ECD (MeOH) λmax (Δε): 233 (−22.78), 370 (+3.13) nm; 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S35–S40 and Table S3; HRESIMS m/z 501.3001 [M + H]+ (calcd for C33H41O4, 501.2999) (Supporting Information Fig. S41).
Cleistoperone E (5): yellow oil; −107.0 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 200 (3.45), 234 (3.02), 298 (3.00), 368 (3.05) nm (Supporting Information Fig. S42); IR 3478, 3411, 2933, 1690, 1634, 1577, 1466, 1424, 1212, 1181, 1127, 764, 623 cm−1 (Supporting Information Fig. S43); ECD (MeOH) λmax (Δε): 233 (−19.95), 263 (−9.33), 334 (−4.12), 356 (+10.12) nm; 1H (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Supporting Information Figs. S44–S49 and Table S3; HRESIMS m/z 501.3002 [M + H]+ (calcd for C33H41O4, 501.2999) (Supporting Information Fig. S50).
Cleistoperone F (6): yellow oil; −69.5 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.53), 231 (3.03), 304 (3.01), 378 (3.36) nm (Supporting Information Fig. S51); IR 3405, 2961, 2927, 2857, 1713, 1639, 1617, 1563, 1458, 1407, 1051, 988, 623 cm−1 (Supporting Information Fig. S52); ECD (MeOH) λmax (Δε): 207 (−14.49), 289 (+8.33), 329 (−1.63), 381 (+4.02) nm; 1H (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Supporting Information Figs. S53–S58 and Table S4; HRESIMS m/z 501.3004 [M + H]+ (calcd for C33H41O4, 501.2999) (Supporting Information Fig. S59).
Cleistoperone G (7): yellow oil; +63.5 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.40), 234 (2.99), 296 (2.97), 375 (3.14) nm (Supporting Information Fig. S60); IR 3487, 3417, 1729, 1639, 1614, 1563, 1455, 1404, 1172, 912, 632 cm−1 (Supporting Information Fig. S61); ECD (MeOH) λmax (Δε): 207 (−20.08), 289 (+4.39), 325 (−0.87), 381 (+2.60) nm; 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S62–S67 and Table S4; HRESIMS m/z 497.2684 [M + H]+ (calcd for C33H37O4, 497.2686) (Supporting Information Fig. S68).
Cleistoperone H (8): yellow oil; ±0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.45), 234 (3.12), 300 (3.05), 375 (3.30) nm (Supporting Information Fig. S69); IR 3481, 3417, 2927, 2867, 1642, 1614, 1563, 1453, 1413, 1172, 1107, 620, 479 cm−1 (Supporting Information Fig. S70); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S71–S76 and Table S5; HRESIMS m/z 501.2995 [M + H]+ (calcd for C33H41O4, 501.2999) (Supporting Information Fig. S77).
(+)-8: +120.0 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 207 (+22.53), 236 (+21.36), 352 (−2.35) nm.
(−)-8: −121.2 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 206 (−33.22), 235 (−29.19), 350 (+3.08) nm.
Cleistoperone I (9): yellow oil; ±0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.45), 344 (3.17) nm (Supporting Information Fig. S78); IR 3417, 2978, 2927, 2864, 1659, 1619, 1511, 1441, 1186, 1093, 1068, 881, 756 cm−1 (Supporting Information Fig. S79); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S80–S85 and Table S5; HRESIMS m/z 501.2997 [M + H]+ (calcd for C33H41O4, 501.2999) (Supporting Information Fig. S86).
(+)-9: +98.5 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 213 (+27.55), 380 (+2.33) nm.
(−)-9: −99.8 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 213 (−30.80), 382 (−2.31) nm.
Cleistoperone J (10): yellow oil; +24.4 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.51), 233 (3.11), 302 (3.08), 376 (3.43) nm (Supporting Information Fig. S87); IR 3484, 3411, 2912, 1735, 1637, 1617, 1554, 1195, 1156, 906, 835, 626 cm−1 (Supporting Information Fig. S88); ECD (MeOH) λmax (Δε): 232 (+20.90), 290 (+4.80) nm; 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S89–S94 and Table S6; HRESIMS m/z 501.3006 [M + H]+ (calcd for C33H41O4: 501.2999) (Supporting Information Fig. S95).
Cleistoperone K (11): yellow oil; −27.6 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.49), 233 (3.11), 300 (3.07), 375 (3.43) nm (Supporting Information Fig. S96); IR 3420, 2930, 1679, 1619, 1583, 1359, 1186, 1027, 926, 858, 730, 956, 609 cm−1 (Supporting Information Fig. S97); ECD (MeOH) λmax (Δε): 236 (−9.50), 292 (−3.19) nm; 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S98–S103 and Table S6; HRESIMS m/z 501.3001 [M + H]+ (calcd for C33H41O4: 501.2999) (Supporting Information Fig. S104).
Cleistoperone L (12): yellow oil; +65.6 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 200 (3.21), 346 (3.01) nm (Supporting Information Fig. S105); IR 3481, 3417, 2946, 2930, 1653, 1610, 1511, 1466, 1444, 1198, 980, 892, 629 cm−1 (Supporting Information Fig. S106); ECD (MeOH) λmax (Δε): 213 (+23.27), 380 (+2.93) nm; 1H (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Supporting Information Figs. S107–S112 and Table S7; HRESIMS m/z 501.3004 [M + H]+ (calcd for C33H41O4: 501.2999) (Supporting Information Fig. S113).
Cleistoperone M (13): yellow oil; −63.6 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 200 (3.40), 344 (3.14) nm (Supporting Information Fig. S114); IR 3484, 3414, 2981, 2913, 1637, 1617, 1574, 1455, 1381, 1078, 756, 629, 479 cm−1 (Supporting Information Fig. S115); ECD (MeOH) λmax (Δε): 217 (−7.05), 385 (−1.76) nm; 1H (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Supporting Information Figs. S116–S121 and Table S7; HRESIMS m/z 501.3008 [M + H]+ (calcd for C33H41O4: 501.2999) (Supporting Information Fig. S122).
Cleistoperone N (14): yellow block-shaped crystals (MeOH/MeCN, 6:4); mp 167–168 °C; −91.4 (c 0.40, MeOH); UV (MeOH) λmax (log ε) 200 (3.36), 348 (3.04) nm (Supporting Information Fig. S123); IR 3481, 3411, 2969, 2924, 1707, 1617, 1568, 1511, 1455, 1384, 1073, 623, 470 cm−1 (Supporting Information Fig. S124); 1H (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Supporting Information Figs. S125–S130 and Table S8; HRESIMS m/z 501.3001 [M + H]+ (calcd for C33H41O4: 501.2999) (Supporting Information Fig. S131).
Cleistoperone O (15): yellow oil; +98.1 (c 0.40, MeOH); UV (MeOH) λmax (log ε) 200 (3.45), 347 (3.16) nm (Supporting Information Fig. S132); IR 3478, 3414, 2961, 2930, 1701, 1637, 1619, 1565, 1540, 1453, 1419, 1121, 875, 832 cm−1 (Supporting Information Fig. S133); ECD (MeOH) λmax (Δε): 206 (+48.80), 233 (−6.87), 342 (+18.65) nm; 1H (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Supporting Information Figs. S134–S139 and Table S8; HRESIMS m/z 501.3002 [M + H]+ (calcd for C33H41O5: 501.2999) (Supporting Information Fig. S140).
Cleistoperone P (16): yellow needle-shaped crystals (MeOH/CH2Cl2/H2O, 7:2:1); mp 185–186 °C; −33.4 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.13), 233 (2.84), 300 (2.80), 378 (3.09) nm (Supporting Information Fig. S141); IR 3481, 3411, 2964, 2927, 1710, 1614, 1514, 1453, 1413, 1158, 1118, 629 cm−1 (Supporting Information Fig. S142); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S143–S148 and Table S9; HRESIMS m/z 433.2371 [M + H]+ (calcd for C28H33O4: 433.2373) (Supporting Information Fig. S149).
(+)-16: +55.5 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 231 (+10.60), 379 (−2.99) nm.
(−)-16: −56.8 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 230 (−10.04), 377 (+2.04) nm.
Cleistoperone Q (17): yellow oil; +73.2 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.44), 234 (3.11), 306 (3.09), 379 (3.43) nm (Supporting Information Fig. S150); IR 3420, 2927, 2859, 1710, 1617, 1514, 1450, 1169, 1113, 974, 915, 700, 620, 490 cm−1 (Supporting Information Fig. S151); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S152–S157 and Table S9; HRESIMS m/z 433.2375 [M + H]+ (calcd for C28H33O4: 433.2373) (Supporting Information Fig. S158).
(+)-17: +78.5 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 208 (−20.60), 291 (+9.70), 381 (+7.78) nm.
(−)-17: −77.5 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 210 (+20.77), 288 (−10.20), 382 (−6.31) nm.
Cleistoperone R (18): yellow plate-shaped crystals (MeCN/CH2Cl2, 9:1); mp 213–214 °C; ±0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 200 (3.27), 233 (3.06), 300 (3.00), 374 (3.06) nm (Supporting Information Fig. S159); IR 3476, 3414, 2958, 2930, 1637, 1614, 1565, 1514, 1447, 1195, 1025, 629 cm−1 (Supporting Information Fig. S160); 1H (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data see Supporting Information Figs. S161–S166 and Table S9; HRESIMS m/z 435.2357 [M + H]+ (calcd for C28H35O4, 435.2350) (Supporting Information Fig. S167).
(+)-18: +105.0 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 203 (+22.52), 279 (−5.12), 318 (+2.99), 355 (−2.17) nm.
(−)-18: −106.6 (c 0.10, MeOH); ECD (MeOH) λmax (Δε): 203 (−25.02), 280 (+5.80), 319 (−3.26), 358 (+2.57) nm.
4.5. X-ray crystallographic analysis
Single-crystal diffraction data were collected via an Oxford Cryo stream system on a XtaLAB PRO MM007-DW diffractometer system equipped with a RA-Micro7HF-MR-DW(Cu/Mo) X-ray generator and Pilatus3R-200K-A detector (Rigaku, Japan, Cu Kα, λ = 1.54178 Å). The unit cell and data reduction were determined using CrysAlisPro software. The numerical absorption corrections were applied using the program of ABSCOR. As we previously described42, the structures were solved by direct methods (ShelXS in Olex2 1.2) and refined by full-matrix least-squares method in the SHELX-2013 program package. In the structure refinements, all non-H atoms were refined anisotropically. The positions of the H-atoms were calculated geometrically with riding models (Supporting Information Figs. S168–S178 and Tables S10–S15). The crystallographic data reported in this study were deposited in the Cambridge Crystallographic Data Centre under accession numbers CCDC 2333986–2333991, respectively.
4.6. Quantum chemical calculations
The 3D molecular structures of compounds 1–13 and 15–18 were built and energy-minimized with ChemBio3D Ultra 11.0 Suite (Cambridge Soft, USA) using MM2 force field. The random conformational searches were performed by SYBYL X 2.1.1 program using MMFF94s molecular force field, with an energy cutoff of 10 kcal/mol to the global minima. The obtained conformers were subsequently optimized by using Gaussion09 software43 at the B3LYP/6-31+G(d) level in gas phase. The optimized stable conformers with an energy window of 3 kcal/mol were selected for further NMR or ECD calculations. The NMR computations were performed at the mPW1PW91/6-31+G(d,p) level with CHCl3 as the solvent. The ECD curves were predicted at the B3LYP/6-31+G(d) level in MeOH. The overall calculated NMR and ECD data were all weighted by Boltzmann distribution, and were subsequently compared with the experimental ones (Supporting Information Figs. S179–S195 and Tables S16–S49). The ECD curves were produced by SpecDis 1.71 software44.
4.7. Biological assay
4.7.1. Cells, virus, and antiviral compounds
The human epithelial type 2 cells (HEp-2) were obtained from American Type Culture Collection (ATCC) and grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). Human respiratory syncytial virus, strain A2 (ATCC, VR-1540), was propagated in HEp-2 cells and titrated by plaque assay45. Ribavirin was purchased from Sigma–Aldrich. The purity of CPTAs was more than 98%, as determined by HPLC analysis. All the tested compounds were dissolved in DMSO at 50 mmol/L, stored at −20 °C, and diluted to final concentrations with cell culture medium.
4.7.2. Time of addition assay
HEp-2 cells were seeded in 96-well plates one day prior to RSV A2 infection. At 0, 2, 6, 12, 18, or 30 h postinfection, the cell culture medium was discarded and replaced with fresh medium containing 1 (10 μmol/L). At 40 h after the viral infection, the viral titers in the cells were determined as previously described20.
4.7.3. Western blot assay
HEp-2 cells were seeded in 6-well plates and incubated overnight. The cells were then infected with RSV A2 in the presence or absence of 1. At 24 h postinfection, the cells were washed with ice-cold PBS and lysed on ice using RIPA buffer with phenylmethylsulfonyl fluoride (PMSF) and phosphatase inhibitor cocktails. The supernatants of cell lysates were collected after centrifuging for 15 min at 4 °C. The protein concentration of each cell lysate was determined using a Pierce BCA protein assay kit. The proteins in cell lysates were reduced and denatured by boiling in a loading buffer at 95 °C for 5 min and then transferred to the PVDF membrane by SDS-PAGE. The membranes were blocked with 5% bovine serum (BSA) in PBST (PBS/0.1% Tween-20) and then incubated with specific protein antibodies including anti-Akt [catalog no. 4685S; Cell Signaling Technology (CST), USA], anti-phospo Akt (catalog No. 66444-1; Proteintech, USA), anti-mTOR (catalog No. 2983T; CST), anti-phospho mTOR (catalog No. 5536T; CST), anti-phospho p70S6K (catalog No. 9208T; CST), anti-4E-BP1 (catalog No. 60246; Proteintech), anti-phospho 4E-BP1 (catalog No. 2855T; CST), anti-eIF4E (catalog No. 101113; SinoBiological, CHN), and anti-GAPDH (catalog No. 5174; CST). After washing thrice with PBST, the specified primary antibodies were stained with respective horseradish peroxidase (HRP)-conjugated secondary antibodies. Finally, the membranes were incubated with ECL Reagent and imaged with a chemiluminescence imaging system (Amersham Imager 600, GE Healthcare).
4.7.4. Immunofluorescence assay
HEp-2 cells were seeded into 96-well plates and incubated for 24 h. The cells were then inoculated with RSV A2 and treated with ribavirin or tested compounds. At 48 h postinfection (h.p.i.), the cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 in PBS for 10 min, followed by washing thrice with PBS. The cells were then blocked with 4% bovine serum albumin (BSA) in PBS for 30 min at room temperature (RT). The specific antibodies against RSV fusion glycoprotein (catalog No. ab94968; Abcam, USA) or cellular 4E-BP1 (catalog No. 60246, Proteintech, USA) and eIF4E (catalog No. 101113, SinoBiological, CHN) were added to the cells and incubated with the cells for 2 h at RT. After washing with PBS, the cells were stained with DyLight 594-conjugated secondary antibody (catalog No. 35510; Thermo Fisher Scientific, USA) for 1 h at RT. The cell nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI). Finally, the cells were photographed under a confocal laser scanning microscope (Zeiss LSM800).
Acknowledgments
This work was financially supported by the National Key R&D Program of China (No. 2023YFC3503902, China), the National Natural Science Foundation of China (Nos. 82293681(82293680), 82321004, 82204234, and 82273822, China), the Guangdong Basic and Applied Basic Research Foundation (Nos. 2022B1515120015 and 2021A1515111021, China), the Guangdong Major Project of Basic and Applied Basic Research (No. 2023B0303000026, China), the Guangdong-Hong Kong-Macau Universities Joint Laboratory for the Internationalization of Traditional Chinese Medicine (No. 2023LSYS002, China), the Guangzhou Key Laboratory of Traditional Chinese Medicine & Disease Susceptibility (No. 2024A03J090, China), and the Science and Technology Projects in Guangzhou (No. 202102070001, China). This work was also supported by the high-performance computing platform of Jinan University.
Author contributions
Jianguo Song: Conceptualization, Investigation, Funding acquisition, Writing – original draft, Writing – review & editing. Ruili Huang: Investigation, Writing – review & editing. Jialiao Cai: Investigation. Zhenlong Wu: Funding acquisition, Investigation, Visualization, Writing – review & editing. Lijun Hu: Investigation, Writing – review & editing. Wanyang Sun: Investigation, Writing – review & editing. Xiaojun Huang: Data curation, Formal analysis, Validation. Rongrong He: Investigation, Funding acquisition. Wei Tang: Investigation, Validation, Writing – review & editing. Wencai Ye: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. Ying Wang: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2024.04.031.
Contributor Information
Wei Tang, Email: tangv163@163.com.
Wencai Ye, Email: chywc@aliyun.com.
Ying Wang, Email: wangying_cpu@163.com.
Appendix A. Supporting information
The following is the Supporting information to this article:
References
- 1.Bouslimani A., Sanchez L.M., Garg N., Dorrestein P.C. Mass spectrometry of natural products: current, emerging and future technologies. Nat Prod Rep. 2014;31:718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krug D., Muller R. Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep. 2014;31:768–783. doi: 10.1039/c3np70127a. [DOI] [PubMed] [Google Scholar]
- 3.Yu Y., Yao C., Guo D. Insight into chemical basis of traditional Chinese medicine based on the state-of-the-art techniques of liquid chromatography–mass spectrometry. Acta Pharm Sin B. 2021;11:1469–1492. doi: 10.1016/j.apsb.2021.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox Ramos A.E., Evanno L., Poupon E., Champy P., Beniddir M.A. Natural products targeting strategies involving molecular networking: different manners, one goal. Nat Prod Rep. 2019;36:960–980. doi: 10.1039/c9np00006b. [DOI] [PubMed] [Google Scholar]
- 5.Kang K.B., Park E.J., da Silva R.R., Kim H.W., Dorrestein P.C., Sung S.H. Targeted isolation of neuroprotective dicoumaroyl neolignans and lignans from Sageretia theezans using in silico molecular network annotation propagation-based dereplication. J Nat Prod. 2018;81:1819–1828. doi: 10.1021/acs.jnatprod.8b00292. [DOI] [PubMed] [Google Scholar]
- 6.Allard P.M., Peresse T., Bisson J., Gindro K., Marcourt L., Pham V.C., et al. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal Chem. 2016;88:3317–3323. doi: 10.1021/acs.analchem.5b04804. [DOI] [PubMed] [Google Scholar]
- 7.Esposito M., Nothias L.F., Retailleau P., Costa J., Roussi F., Neyts J., et al. Isolation of premyrsinane, myrsinane, and tigliane diterpenoids from Euphorbia pithyusa using a chikungunya virus cell-based assay and analogue annotation by molecular networking. J Nat Prod. 2017;80:2051–2059. doi: 10.1021/acs.jnatprod.7b00233. [DOI] [PubMed] [Google Scholar]
- 8.Fox Ramos A.E., Alcover C., Evanno L., Maciuk A., Litaudon M., Duplais C., et al. Revisiting previously investigated plants: a molecular networking-based study of Geissospermum laeve. J Nat Prod. 2017;80:1007–1014. doi: 10.1021/acs.jnatprod.6b01013. [DOI] [PubMed] [Google Scholar]
- 9.Cabral R.S., Allard P.M., Marcourt L., Young M.C.M., Queiroz E.F., Wolfender J.L. Targeted isolation of indolopyridoquinazoline alkaloids from Conchocarpus fontanesianus based on molecular networks. J Nat Prod. 2016;79:2270–2278. doi: 10.1021/acs.jnatprod.6b00379. [DOI] [PubMed] [Google Scholar]
- 10.Ren Y.M., Zhou S.Z., Zhang T., Qian M., Zhang R., Yao S., et al. Targeted isolation of two disesquiterpenoid macrocephadiolides A and B from Ainsliaea macrocephala using a molecular networking-based dereplication strategy. Org Chem Front. 2020;7:1481–1489. [Google Scholar]
- 11.He Q.F., Wu Z.L., Li L., Sun W.Y., Wang G.Y., Jiang R.W., et al. Discovery of neuritogenic Securinega alkaloids from Flueggea suffruticosa by a building blocks-based molecular network strategy. Angew Chem Int Ed. 2021;60:19609–19613. doi: 10.1002/anie.202103878. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Liu F., Jin Q., Li X., Zhan Q., Chen M., et al. Discovery of unusual phloroglucinol-triterpenoid adducts from Leptospermum scoparium and Xanthostemon chrysanthus by building blocks-based molecular networking. Chin Chem Lett. 2024;35 [Google Scholar]
- 13.Li Z.W., Fan C.L., Sun B., Huang L., Wang Z.Q., Huang X.J., et al. Discovery of unusual ajmaline-macroline type bisindole alkaloids from Alstonia macrophylla by building blocks-based molecular networking. Chem Eur J. 2024;30 doi: 10.1002/chem.202303519. [DOI] [PubMed] [Google Scholar]
- 14.Zheng X., Gao L., Wang L., Liang C., Wang B., Liu Y., et al. Discovery of ziresovir as a potent, selective, and orally bioavailable respiratory syncytial virus fusion protein inhibitor. J Med Chem. 2019;62:6003–6014. doi: 10.1021/acs.jmedchem.9b00654. [DOI] [PubMed] [Google Scholar]
- 15.Cockerill G.S., Good J.A.D., Mathews N. State of the art in respiratory syncytial virus drug discovery and development. J Med Chem. 2019;62:3206–3227. doi: 10.1021/acs.jmedchem.8b01361. [DOI] [PubMed] [Google Scholar]
- 16.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 17.Luo Z., Yin F., Wang Y., Kong L. Progress in approved drugs from natural product resources. Chin J Nat Med. 2024;22:195–211. doi: 10.1016/S1875-5364(24)60582-0. [DOI] [PubMed] [Google Scholar]
- 18.Guo Z. The modification of natural products for medical use. Acta Pharm Sin B. 2017;7:119–136. doi: 10.1016/j.apsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y., Ouyang Z., Du H., Wang M., Wang J., Sun H., et al. New opportunities and challenges of natural products research: when target identification meets single-cell multiomics. Acta Pharm Sin B. 2022;12:4011–4039. doi: 10.1016/j.apsb.2022.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W., Li M., Liu Y., Liang N., Yang Z., Zhao Y., et al. Small molecule inhibits respiratory syncytial virus entry and infection by blocking the interaction of the viral fusion protein with the cell membrane. FASEB J. 2019;33:4287–4299. doi: 10.1096/fj.201800579R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J., Huang H., Li B., Li H., Zhao Y., Li Y., et al. Identification of cytochrome c oxidase subunit 4 isoform 1 as a positive regulator of influenza virus replication. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.862205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan L., Wang Y., Liang N., Huang X.J., Li M.M., Fan C.L., et al. Chemical constituents from the roots and stems of Erycibe obtusifolia and their in vitro antiviral activity. Planta Med. 2013;79:1558–1564. doi: 10.1055/s-0033-1350804. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Chen M., Zhang J., Zhang X.L., Huang X.J., Wu X., et al. Flavone C-glycosides from the leaves of Lophatherum gracile and their in vitro antiviral activity. Planta Med. 2012;78:46–51. doi: 10.1055/s-0031-1280128. [DOI] [PubMed] [Google Scholar]
- 24.Pham G.N., Nguyen T.T.T., Nguyen-Ngoc H. Ethnopharmacology, phytochemistry, and pharmacology of Syzygium nervosum. Evid-Based Compl Alt. 2020;2020 doi: 10.1155/2020/8263670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha T.K.Q., Dao T.T., Nguyen N.H., Kim J., Kim E., Cho T.O., et al. Antiviral phenolics from the leaves of Cleistocalyx operculatus. Fitoterapia. 2016;110:135–141. doi: 10.1016/j.fitote.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Wang C., Wu P., Tian S., Xue J., Xu L., Li H., et al. Bioactive pentacyclic triterpenoids from the leaves of Cleistocalyx operculatus. J Nat Prod. 2016;73:2912–2923. doi: 10.1021/acs.jnatprod.6b00715. [DOI] [PubMed] [Google Scholar]
- 27.Dao T.T., Tung B.T., Nguyen P.H., Thuong P.T., Yoo S.S., Kim E.H., et al. C-Methylated flavonoids from Cleistocalyx operculatus and their inhibitory effects on novel influenza A (H1N1) neuraminidase. J Nat Prod. 2010;73:1636–1642. doi: 10.1021/np1002753. [DOI] [PubMed] [Google Scholar]
- 28.Su J.C., Wang S., Cheng W., Huang X.J., Li M.M., Jiang R.W., et al. Phloroglucinol derivatives with unusual skeletons from Cleistocalyx operculatus and their in vitro antiviral activity. J Org Chem. 2018;83:8522–8532. doi: 10.1021/acs.joc.8b01050. [DOI] [PubMed] [Google Scholar]
- 29.Song J.G., Su J.C., Song Q.Y., Huang R.L., Tang W., Hu L.J., et al. Cleistocaltones A and B, antiviral phloroglucinol-terpenoid adducts from Cleistocalyx operculatus. Org Lett. 2019;21:9579–9583. doi: 10.1021/acs.orglett.9b03743. [DOI] [PubMed] [Google Scholar]
- 30.Song J.G., Liu J.X., Huang R.L., Tang W., Huang X.J., Wang Y., et al. Tautomeric cinnamoylphloroglucinol-monoterpene adducts from Cleistocalyx operculatus and their antiviral activities. J Asian Nat Prod Res. 2024;26:38–51. doi: 10.1080/10286020.2023.2288290. [DOI] [PubMed] [Google Scholar]
- 31.Grimblat N., Zanardi M.M., Sarotti A.M. Beyond DP4: an improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J Org Chem. 2015;80:12526–12534. doi: 10.1021/acs.joc.5b02396. [DOI] [PubMed] [Google Scholar]
- 32.Bertolasi V., Gilli P., Ferretti V., Gilli G. Evidence for resonance-assisted hydrogen bonding. 2. Intercorrelation between crystal structure and spectroscopic parameters in eight intramolecularly hydrogen bonded 1,3-diaryl-1,3-propanedione enols. J Am Chem Soc. 1991;113:4917–4925. [Google Scholar]
- 33.Gilli P., Bertolasi V., Pretto L., Ferretti V., Gilli G. Covalent versus electrostatic nature of the strong hydrogen bond: discrimination among single, double, and asymmetric single-well hydrogen bonds by variable-temperature X-ray crystallographic methods in β-diketone enol RAHB systems. J Am Chem Soc. 2004;126:3845–3855. doi: 10.1021/ja030213z. [DOI] [PubMed] [Google Scholar]
- 34.Bonilla A., Duque C., Garzon C., Takaishi Y., Yamaguchi K., Hara N., et al. Champanones, yellow pigments from the seeds of champa (Campomanesia lineatifolia) Phytochemistry. 2005;66:1736–1740. doi: 10.1016/j.phytochem.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Wang J., Song J.G., Zhong D.L., Duan Z.Z., Peng Z.J., Tang W., et al. Biomimetic synthesis of an antiviral cinnamoylphloroglucinol collection from Cleistocalyx operculatus: a synthetic strategy based on biogenetic building blocks. Angew Chem Int Ed. 2023;62 doi: 10.1002/anie.202312568. [DOI] [PubMed] [Google Scholar]
- 36.Wang P., Li R.J., Liu R.H., Jian K.L., Yang M.H., Yang L., et al. Sarglaperoxides A and B, sesquiterpene-normonoterpene conjugates with a peroxide bridge from the seeds of Sarcandra glabra. Acta Pharm Sin B. 2023;13:754–764. doi: 10.1021/acs.orglett.6b00112. [DOI] [PubMed] [Google Scholar]
- 37.Wang L., Xia G., Xia H., Wei X., Wang Y., Lin S. (+)/(−)-Yanhusamides A–C, three pairs of unprecedented benzylisoquinoline-pyrrole hetero-dimeric alkaloid enantiomers from Corydalis yanhusuo. Org Lett. 2016;18:832–835. doi: 10.1016/j.apsb.2022.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo S.L., Hu L.J., Huang X.J., Su J.C., Shao X.H., Wang L., et al. Discovery and biomimetic synthesis of a phloroglucinol-terpene adduct collection from Baeckea frutescens and its biogenetic origin insight. Chem Eur J. 2020;26:11104–11108. doi: 10.1002/chem.202001111. [DOI] [PubMed] [Google Scholar]
- 39.Hou J.Q., Guo C., Zhao J.J., He Q.W., Zhang B.B., Wang H. Frutescone A–G, tasmanone-based meroterpenoids from the aerial parts of Baeckea frutescens. J Org Chem. 2017;82:1448–1457. doi: 10.1021/acs.joc.6b02643. [DOI] [PubMed] [Google Scholar]
- 40.Yang X.W., Li Y.P., Su J., Ma W.G., Xu G. Hyperjapones A–E, terpenoid polymethylated acylphloroglucinols from Hypericum japonicum. Org Lett. 2016;18:1876–1879. doi: 10.1021/acs.orglett.6b00650. [DOI] [PubMed] [Google Scholar]
- 41.Murray A.F., Moore A.J., Munafo J.P. Key odorants from the American matsutake, Tricholoma magnivelare. J Agric Food Chem. 2020;68:9768–9775. doi: 10.1021/acs.jafc.0c03372. [DOI] [PubMed] [Google Scholar]
- 42.Song J.G., Zheng J., Wei R.J., Huang Y.L., Jiang J., Ning G.H., et al. Crystalline mate for structure elucidation of organic molecules. Chem. 2024;10:924–937. [Google Scholar]
- 43.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., et al. Gaussian, Inc; Wallingford CT: 2013. Gaussian 09, revision E.01. [Google Scholar]
- 44.Bruhn T., Schaumloffel A., Hemberger Y., Pescitelli G. 2017. SpecDis version 1.71. Berlin, Germany. [Google Scholar]
- 45.Tang W., Li Y., Song Q., Wang Z., Li M., Zhang Q., et al. Mechanism of cross-resistance to fusion inhibitors conferred by the K394R mutation in respiratory syncytial virus fusion protein. J Virol. 2021;95 doi: 10.1128/JVI.01205-21. e01205-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.