Abstract
Background
The genomes within organelles are crucial for physiological functions such as respiration and photosynthesis and may also contribute to environmental adaptation. However, the limited availability of genetic resources, particularly mitochondrial genomes, poses significant challenges for in-depth investigations.
Results
Here, we explored various assembly methodologies and successfully reconstructed the complex organelle genomes of two Rhododendron species: Rhododendron nivale subsp. boreale and Rhododendron vialii. The mitogenomes of these species exhibit various conformations, as evidenced by long-reads mapping. Notably, only the mitogenome of R. vialii can be depicted as a singular circular molecule. The plastomes of both species conform to the typical quadripartite structure but exhibit elongated inverted repeat (IR) regions. Compared to the high similarity between plastomes, the mitogenomes display more obvious differences in structure, repeat sequences, and codon usage. Based on the analysis of 58 organelle genomes from angiosperms inhabiting various altitudes, we inferred the genetic adaptations associated with high-altitude environments. Phylogenetic analysis revealed partial inconsistencies between plastome- and mitogenome-derived phylogenies. Additionally, evolutionary lineage was determined to exert a greater influence on codon usage than altitude. Importantly, genes such as atp4, atp9, mttB, and clpP exhibited signs of positive selection in several high-altitude species, suggesting a potential link to alpine adaptation.
Conclusions
We tested the effectiveness of different organelle assembly methods for dealing with complex genomes, while also providing and validating high-quality organelle genomes of two Rhododendron species. Additionally, we hypothesized potential strategies for high-altitude adaptation of organelles. These findings offer a reference for the assembly of complex organelle genomes, while also providing new insights and valuable resources for understanding their adaptive evolution patterns.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05761-7.
Keywords: Organelle genome, Assembly methodology, Phylogeny, High-altitude adaptation
Background
Mitochondria and plastids, organelles with genomes of endosymbiotic origin, are key to regulating photosynthesis and respiration [1]. They possess their own independent genomes, namely mitogenome and plastome, which are crucial for organelle function and overall life processes [2]. Since their emergence, mitochondria have undergone rapid diversification [3]. The mitogenomes of most land plants exhibit significant variation due to recombination events mediated by repetitive sequences, allowing them to exist in various structural forms, including circular, linear, branched, multichromosomal, or combinations thereof [4]. In contrast, plastomes in angiosperms are simpler and more conserved [5]. Plastomes are typically smaller and most exhibit a quadripartite circular structure, consisting of a large single copy (LSC) region and a small single copy (SSC) region, separated by a pair of inverted repeats (IRs) [6]. This stable structure makes plastomes relatively easy to obtain, resulting in a greater number of published plastomes compared to mitogenomes. Currently, about 13,000 complete plastomes have been sequenced, compared to only 673 mitogenomes, with only 285 species having both organelle genomes sequenced [1]. This extreme imbalance in sequencing abundance limits our understanding of the evolution and function of plant organelles. With the rapid advancement of sequencing technologies and a growing understanding of plant evolution, it is imperative to swiftly address the quantitative gaps.
As plant adaptation to environmental changes becomes an increasingly critical area of focus in plant science, emerging evidence underscores the significant role of organelles in facilitating this adaptation. In high-altitude environments, plants encounter unique stressors such as low temperatures, high UV radiation, and reduced oxygen levels [7]. Organelles play a crucial role in these adaptations by adjusting their functions to optimize photosynthesis, respiration, and stress responses under such extreme conditions. For example, during cooling phases, cold-tolerant plants exhibit ultrastructural modifications within their organelles as a strategic adaptation to mitigate these stressors [8]. Moreover, molecular variations within plant cytoplasmic genomes often exhibit adaptive significance, as evidenced by phenomena such as positive selection, plastid capture, and nucleocytoplasmic interactions, which contribute to the evolutionary fitness of the species [9]. However, the limited number of protein-coding genes (PCGs) in organelles often leads to the underestimation of significance in plant adaptation [10].
Rhododendron L., the largest genus in the Ericaceae family with over 1,000 species, is a key representative of the highly diverse Sino-Himalayan flora in East Asia, shaped by the heterogeneous topography and climate following the uplift of the Qinghai-Tibet Plateau [11]. It has a wide vertical distribution range, ranging from 300 m to 5400 m above sea level. Additionally, due to its remarkable adaptability and colorful corolla, Rhododendron possesses high horticultural value. Although phylogenetic studies of plastome PCGs across 161 species have been performed, the structural variations in the plastome of Rhododendron remain unclear, mainly due to challenges in genome assembly [12, 13]. At present, only two mitochondrial scaffolds have been published, R. × pulchrum and R. simsii [14, 15]. For plastome, about 30 complete genomes have been published on NCBI (https://www.ncbi.nlm.nih.gov/), ranging in length from 146,941 bp to 230,777 bp. In the face of a large genus of more than 1,000 species, this is far from sufficient. Therefore, providing more complete and accurate organelle genomes is of great significance for understanding the adaptive evolution of Rhododendron and revealing the uplifting history of the Qinghai-Tibet Plateau.
Organelles hold a significant position in the biological activities of life, and exploring their evolutionary patterns and adaptive strategies enhances our understanding of the driving forces behind organelle differentiation [16]. In addition, the Rhododendron genus, known for its extreme adaptability and genomic diversity, is ideal for comparative genomic research. However, the complex organelle genomes of Rhododendron species present significant challenges. Here, we evaluated various assembly methods to identify the best strategies for reconstructing Rhododendron organelle genomes. Based on the gap-free genomes for R. nivale subsp. boreale and R. vialii, we conducted a comprehensive analysis of their structures, repetitive sequences, RNA editing sites, codon usage, and homologous sequences. To investigate the adaptive evolution of organelle genomes across altitudes, we collected organelle genomes from 58 species at different altitudes. These genomes were used to explore phylogenetic relationships, altitude-associated codon usage patterns, and positive selection genes in high-altitude environments. These findings will serve as a reference for assembling complex organelle genomes and provide a foundation for further research into the adaptive evolution of organelles.
Materials and methods
Plant materials and sequencing
Fresh leaves of R. nivale subsp. boreale were collected from Baima Mountain, Dêqên County, Yunnan Province, China (99°4′13′′E, 28°20′24′′N, alt. 4287.5 m). The plant sample was identified by Zhen-Yu Lyu. The voucher specimens were preserved in the Herbarium, Yunnan University (YUKU). Long-read libraries were constructed and sequenced using the PacBio Sequel II sequencing platform. For short reads, DNA libraries were constructed and sequenced on the BGI DNBseq sequencing platform. In addition, details of other genome data used are listed in Table S1. The raw data used for the genome reassembly of R. vialii and R. molle were obtained from existing studies (Table S2) [17, 18].
Organelle genome assembly and annotation
To obtain the complete organelle genome of Rhododendron, we tested several organelle assembly software to test their assembly efficiency organelle genome. Three species were tested, R. nivale subsp. boreale, R. vialii and R. molle. The data sets for R. nivale subsp. boreale and R. vialii contained HiFi reads, DNB reads, while those for R. molle were Nanopore (ONT) reads, and Illumina reads. Firstly, 10 ~ 20× short-read datasets were randomly extracted. Then we tested GetOrganelle, NOVOPlasty, ptGAUL, Flye + Unicycler, PMAT, Oatk respectively. (1) GetOrganelle v1.7.7.0 [19]. We ran based on next-generation sequencing (NGS) data (DNB and Illumina) from three Rhododendron species using recommended parameters for the plastome and mitogenome. (2) NOVOPlasty v4.3.1 [20]. NOVOPlasty was employed to assemble plastomes using short reads from Illumina and DNB sequencing. The complete plastome sequence of R. griersonianum was used as a seed. (3) Flye v2.9.3 [21] + Unicycler v0.5.0 [22]. Flye and Unicycler were commonly used plant mitogenome assembly pipeline. First, we used Flye for initial assembly based on HIFI reads and ONT reads. Next, BLASTn program was used to search for mitochondrial contigs based on the mitogenome of R. × pulchrum [14]. Then, we filtered short reads by mapping to the mitochondrial contigs with BWA v0.7.17-r1188 [23]. Finally, Unicycler was used for hybrid assembly based on short reads and long reads. (4) PMAT v1.5.1 [24]. Based on long reads, PMAT was used to assemble plastomes and mitogenomes with default parameters. (5) ptGAUL v1.0.5 [25]. ptGAUL was used for assembly of the plastome and we set coverage at 150. (6) Oatk v1.0 (https://github.com/c-zhou/oatk). Oatk was a tool specifically crafted for the de novo assembly of intricate plant organelle genomes using PacBio HiFi data. We assembled two organelle genomes using Oatk, with coverage set at 150. Detailed assembly information was provided in the Table S3. The final organelle genomes were generated after adjustment using Bandage v0.8.1 [26]. Geseq [27] was used to annotate organelle genomes. Manually check and correct gene boundaries using the Apollo v2.7.0 [28]. The organelle genome maps were created using OGDRAW [29].
Structure analysis
Our assembly results in a multi-branch structure, especially R. nivale subsp. boreale, which cannot be restored to a circle. We hypothesize that the mitogenome may have a complex conformation mediated by repeated sequences, like Abelmoschus esculentus [30]. To detect these configurations, we mapped long-read reads back to all paths using minimap2 v2.26-r1175 [31]. About 1,000 bp is taken from each side of the multipath overlap region, and only when at least one read length completely covers the region, we consider it a possible conformation. The Integrative Genomics Viewer (IGV) v2.17.0 [32] is used to visualize genome comparison results. The same strategy was used to verify plastome structure.
RNA editing site prediction
We used Deepred-Mt [33] to predict RNA editing sites of mitogenome. Deepred-Mt was a neural network able to predict C-to-U editing sites in angiosperm mitochondria. Sites with scores greater than 0.9 were considered to be trusted RNA editing sites.
Repeat sequence analysis
Simple sequence repeats (SSRs) were identified using MISA v2.1 [34] with the following parameters: “1–10 2–5 3–4 4 − 3 5 − 3 6 − 3”. Tandem Repeats Finder (TRF) v4.09 [35] program was used to detect tandem repeats. Dispersed_repeats were identified using REPuter [36] with the minimum repeat size was set to 30 bp and hamming distance to 3, and four duplicate types were detected, including F (forward), P (palindrome), R (reverse), and C (complement).
Homologous fragment and collinearity analysis
To identify chloroplast DNA fragments in the mitogenome. We employed BLASTn to search the mitochondrial and plastomes of R. nivale subsp. boreale and R. vialii, respectively. The collinear analysis compared the mitogenomes of three species (two Rhododendron species and Vaccinium macrocarpon) with BLASTn. The E set to 1e-5 and the identity threshold of 70. The result was visualized using NGenomeSyn v1.41 [37] and Circos v0.69 [38].
Phylogenetic analysis
In order to understand the high-altitude adaptation mechanism of organelle genome. We screened 106 angiosperm families for potential altitudinal variation and selected 58 species from 11 families, each possessing both plastomes and mitogenomes, that exhibit significant differences in altitude. They almost represent all available organelle genomes from published data that can be used to explore adaptation to altitudinal differences. Oryza sativa was selected as the outgroup. In total, 59 species were used to construct the phylogenetic tree. In total, 32 shared mitochondrial PCGs (24 core genes and 8 variable genes) and 77 shared plastome PCGs were extracted using Phylosuite v1.2.2 (Table S4) [39] and aligned using MAFFT v7.520 [40]. Additionally, trimAl v1.4 [41] was used for the automated removal of spurious sequences or poorly aligned regions. The concatenation methods were used to reconstruct phylogeny. The IQtree v.2.2.2.6 [42] was used to was used to construct plastome- and mitogenome-based phylogenetic trees, respectively. The parameters were set to “-bb 1000 -m MFP”. The iTOL (Interactive Tree Of Life) v6.9 [43] was used to visualize phylogenetic trees.
Codon usage analysis
Codon usage was influenced by multiple factors: mutational bias, selection, and GC-biased gene diversity. We aimed to explore the impact of high-altitude plants on codon usage. CodonW v1.4.4 (https://codonw.sourceforge.net/) was used to calculate relative synonymous codon usage (RSCU) values, effective number of codons (ENc), and GC content at the third position of codons (GC3s). We used the Wilcoxon Signed-Rank Test to assess differences in median values among multiple independent samples. The level of significance was set at P value < 0.05. Moreover, we also explored the frequency of codon usage from different habitats and evolutionary lineages, based on analysis using EMBOSS v6.6.0 [44]. Codon frequencies are divided into absolute codon usage frequency (absCUFs) and synonymous codon usage frequency (synCUFs) [45]. The absCUFs were normalized based on the total number of codons used, while synCUFs was normalized based on the synonymous codons. Principal Component Analysis (PCA) analysis was performed using “prcomp” fuction and visualized using the ggplot2 package [46] in R v4.3.2 [47].
Positive selection analysis
We used MACSE pipeline v2.07 [48] to perform the codon-aware alignment of CDSs. Next, we construct phylogenetic trees for each CDS matrix using IQtree v.2.2.2.6 [42] with 1,000 repetitions. These gene trees, along with matrices, were used to perform positive selection analysis via HyPhy v1.2.13 [49] with adaptive Branch-Site Random Effects Likelihood (aBSREL) [50]. Genes with a Ka/Ks ratio > 1 and a P-adjust value < 0.05 were considered to have undergone positive selection on specific branches. Genes that experience positive selection in multiple high-altitude species were defined as potential high-altitude adaptation genes.
Result
Organelle genome assembly and structure analysis of Rhododendron
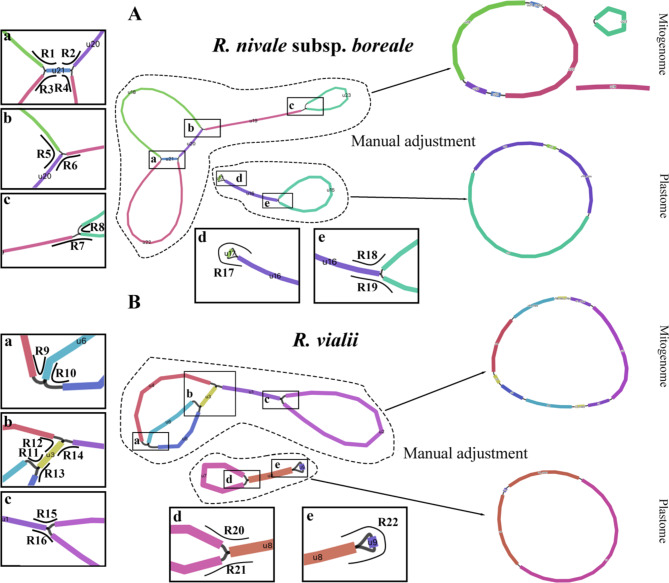
We tested and assembled the mitochondrial and chloroplast genomes of three Rhododendron species: R. nivale subsp. boreale, R. vialii, and R. molle. To achieve this, we used three types of sequencing data: NGS (DNBseq and illumina), and long-read sequencing (ONT and PacBio HiFi) (Table S3). Six approaches were used to test: GetOrganelle (NGS data), NOVOPlasty (NGS data), Unicycler (NGS + long-read sequencing data), ptGAUL (long-read sequencing data), PMAT (long-read sequencing data) and Oatk (HiFi data). Most of the software tested failed to yield correct assembly results (Table S3). Ultimately, we successfully assemble the genomes of R. nivale subsp. boreale and R. vialii, but not R. molle. Specifically, PMAT was effective only for R. nivale subsp. boreale (Fig. S1), while Oatk successfully completed the organelle assemblies for both R. nivale subsp. boreale and R. vialii, providing high-quality sequence graphs. After manually correcting the annotation, we achieved gap-free mitogenomes and plastomes for these two Rhododendron species, which were further confirmed by the uniformity coverage (Figs. 1, S2–S5).
Fig. 1.
The organelle genome structures were visualized by Bandage of R. nivale subsp. boreale (A) and R. vialii (B). The left black box represents a potential region of multiple conformations. R1-R22 represent different hypothetical paths of the mitogenome and plastome. The middle shows drafts of the organelle genome assemblies. The right are the recombination structures of the organelle genomes. Color bands represent contigs
Characters of organelle genomes
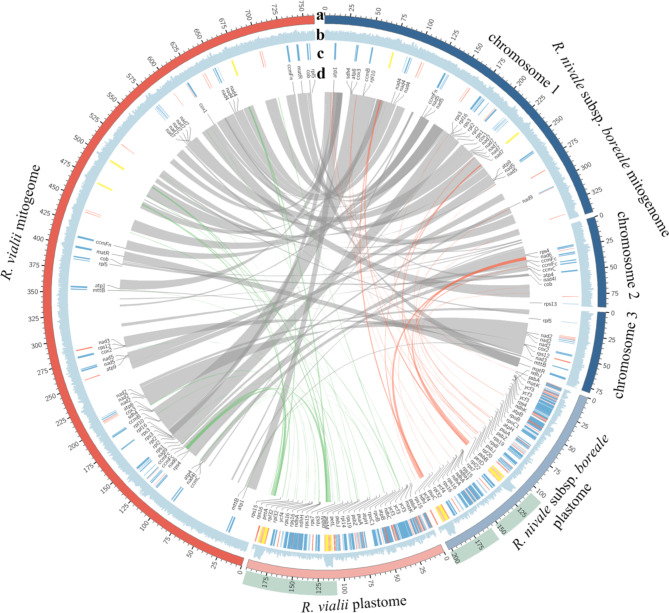
The mitogenome of R. nivale subsp. boreale was divided into three chromosomes: two circular and one linear, with lengths of 342,791 bp, 88,293 bp, and 79,526 bp, respectively, totaling 510,610 bp (Figs. 2 and S6). The average GC content was 46.1%. In contrast, the mitogenome of R. vialii mitogenome was organized into a single circular chromosome with a length of 761,073 bp and an average GC content of 45.9%. We annotated a total of 50 unique genes in the mitogenome of R. nivale subsp. boreale, comprising 30 PCGs, 17 tRNA genes, and three rRNA genes (Table S5). Similarly, the mitogenome of R. vialii contained 51 unique genes, including 31 PCGs, 17 tRNA genes, and three rRNA genes (Table S6).
Fig. 2.
Circos plot represents the organelle genomes of R. nivale subsp. boreale and R. vialii. (a) The length of the organelle genomes; (b) GC density; (c) Genes locations. Blue shows protein-coding genes (PCGs), red shows tRNAs, and yellow shows rRNAs; (d) PCG names and locations. The internal lines represent collinear sequences. The gray lines are homologous fragments between the mitogenomes of the two species. The red lines are homologous fragments between the mitogenome and plastome of R. nivale subsp. boreale. The green lines are homologous fragments between the mitogenome and plastome of R. vialii. The dark green blocks represent the inverted repeat (IR) regions of the plasmid genome
For the plastome, both R. nivale subsp. boreale and R. vialii featured circular genomes and had the tetrad structure, with lengths of 202,882 bp and 195,319 bp, respectively (Figs. 2 and S6). Notably, the plastomes of Rhododendron species had longer inverted repeat (IR) sequences than most angiosperms, with each IR exceeding 43,000 bp. The GC content was 36% for both R. nivale subsp. boreale and R. vialii. Both species were annotated with 111 unique genes, containing 77 PCGs, 30 tRNAs, and four rRNAs (Tables S7-S8).
Structure analysis
Although both R. nivale subsp. boreale and R. vialii belong to the Rhododendron genus, there are significant differences in the structure of their organelle genomes. For the mitogenome, the genomes of these two species may contain multiple potential configurations. To verify these configurations, we extracted sequences of approximately 1,000 bp from each side of various overlapping regions in R. nivale subsp. boreale and R. vialii, encompassing 16 paths (R1-R16; Fig. 1). HiFi reads were used to filter out possible configurations, and a path was considered valid if fully covered by at least one long read. Ultimately, all paths were verified, suggesting that the mitogenomes of both Rhododendron species possess complex structures (Figs. S7-S8). For demonstration purposes, we attempted to restore the mitogenome to a closed loop molecule without branches. However, we only succeeded in R. vialii, while R. nivale subsp. boreale could not be reduced to a circular form (Fig. 1).
An equivalent validation strategy was applied to the plastomes. Verification of the six paths (R17-R22) confirmed the accuracy of our assembly (Figs. S9-S10). The structural variation of the plastome is mainly reflected in the contraction and expansion of IR regions. In the R. nivale subsp. boreale plastome, the length of the IR region was 46,750 bp (Fig. S11). The IRa/LSC junction was located between trnH and trnT, 123 bp from trnH. The IR/SSC junctions were located between rpl32 and ndhF. For R. vialii, the IR region was 43,619 bp in size. The IRa/LSC junction was located within the trnT. Additionally, ndhF was included in the IR region, resulting in an additional copy of ndhF compared to R. nivale subsp. boreale. The complex movement of the IR region boundaries resulted in the SSC region length of R. vialii being only 73 bp, which is significantly smaller than the 2,616 bp of R. nivale subsp. boreale, and devoid of any genes.
Prediction of RNA editing sites
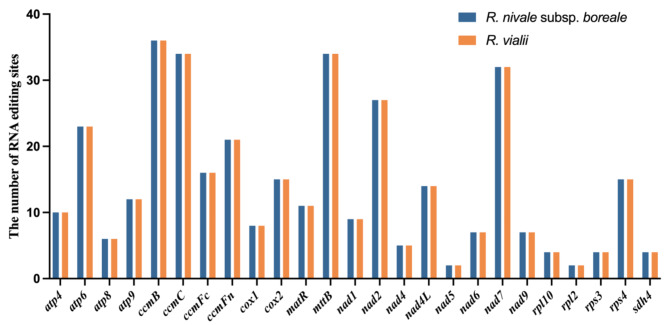
To provide information on possible C-to-U RNA editing in the mitogenomes of Rhododendron species, we predicted potential C-to-U RNA editing sites for R. nivale subsp. boreale and R. vialii. Both species had 358 predicted C-to-U RNA editing sites (Tables S9–S10). Among these, ccmB, ccmC, and mttB contained the most C-to-U RNA editing sites. In contrast, no potential C-to-U RNA editing sites were detected in the genes atp1, cob, cox3, nad3, rpl5, and rps12 in either species (Fig. 3).
Fig. 3.
The distribution of C-to-U RNA editing prediction sites on PCGs for R. nivale subsp. boreale and R. vialii
Repeat sequence analysis
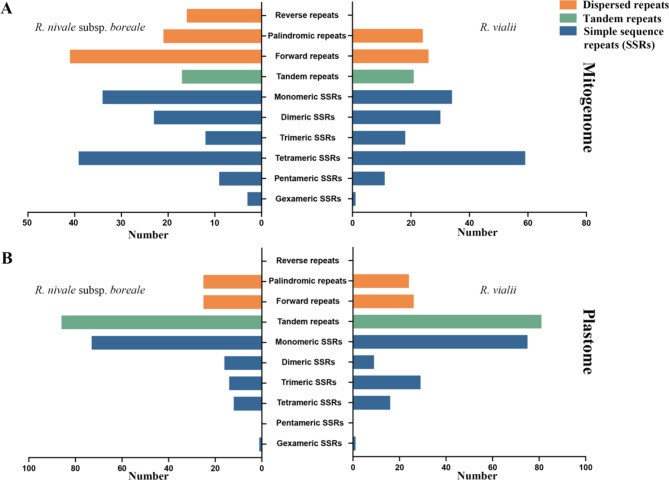
Repetitive sequence analysis was performed for both the mitogenomes and plastomes of R. nivale subsp. boreale and R. vialii. In the mitogenome, SSRs were identified, with R. nivale subsp. boreale having 116 SSRs and R. vialii having 149 SSRs (Tables S11–S12). Six types of SSR were detected, including monomer, dimer, trimer, tetramer, pentamer, and hexamer repeats SSRs, each varying in number. Both species possessed 34 monomeric SSRs and one hexameric SSRs. R. vialii had more dimeric SSRs (28 vs. 22), trimeric SSRs (18 vs. 12), tetrameric SSRs (57 vs. 38), and pentameric SSRs (11 vs. 9) (Fig. 4). We identified 17 tandem repeats in R. nivale subsp. boreale and 27 in R. vialii (Tables S13–S14). These repeats had a similarity of over 81% and lengths ranging from 12 to 42 bp. Additionally, dispersed repeats were detected in both mitogenomes, including palindromic repeats (P), forward repeats (F), and reverse repeats (R) (Tables S15–S16). Notably, 16 R were detected in R. nivale subsp. boreale and none in R. vialii (Fig. 4A).
Fig. 4.
The character of repeats in organelle genomes of R. nivale subsp. boreale and R. vialii. (A) The types and numbers of repeats in the mitogenome; (B) The types and numbers of repeats in the plastome
For the plastomes, R. nivale subsp. boreale possessed 116 SSRs, 86 tandem repeats, and 50 dispersed repeats; while R. vialii had 153 SSRs, 81 tandem repeats, and 50 dispersed repeats (Tables S17–S22). Unlike mitogenome, pentameric SSRs and reverse repeats were not identified in the plastomes (Fig. 4B).
Homologous analysis of mitogenomes and plastomes
Based on the BLAST search, we explored homologous sequences between the mitogenomes and plastomes of R. nivale subsp. boreale and R. vialii. Due to the presence of repeat sequences, the homologous sequences in the mitogenome and plastome do not correspond on a one-to-one basis. In mitogenome of R. nivale subsp. boreale, 55 homologous sequences were identified, with a total length of 12,078 bp, constituting 2.37%. These sequences matched 79 fragments of the plastome, with a total length of 19,269 bp, constituting 9.50%. The sequence identity of these fragments was more than 70%, ranging in length from 31 to 4,162 bp (Table S23). Furthermore, one intact plastome PCG (petL) and eight tRNAs (trnW-CCA, trnN-GUU, trnM-CAU, trnI-GAU, trnI-CAU, trnH-GUG, trnD-GUC, and trnA-UGC) were completely shared between the plastome and mitogenome. Partial shared sequences of 14 additional genes (trnT-GGU, trnI-GAU, trnF-GAA, rrn23, rrn16, rps12, psbL, psbF, psbB, psbA, ndhF, ndhA, atpA, and accD) were also identified.
In the organelle genome of R. vialii, 105 and 92 homologous sequences were identified in the mitogenome and plastome, respectively. The homologous sequence length of the mitogenome was 24,794 bp (3.26%), corresponding to 22,651 bp in the plastome (11.58%). These homologous sequences ranged in length from 32 to 4,150 bp (Table S24). In total, eight genes (petL, trnW-CCA, trnN-GUU, trnM-CAU, trnI-GAU, trnH-GUG, trnD-GUC, and trnA-UGC) were fully shared between the organelle genomes. Additionally, 21 genes (trnI-GAU, trnI-CAU, trnF-GAA, rrn23, rrn16, ycf3, rps12, rpoC2, rbcL, psbL, psbF, psbC, psbB, psbA, ndhF, ndhE, ndhD, ndhA, atpB, atpA, and accD) were partially shared.
Collinearity analysis
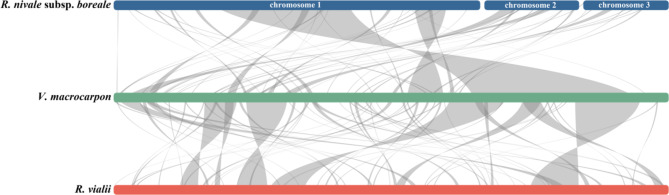
To better elucidate the conservatism and diversity of mitogenome evolution in the Ericaceae, we performed a collinearity analysis of the mitogenome sequences between Rhododendron and Vaccinium, excluding collinear fragments shorter than 300 bp. The results showed that 65 collinear fragments were identified between R. nivale subsp. boreale and R. vialii, spanning 403,872 bp (79.1%) and 476,594 bp (62.6%) of their respective mitogenomes (Fig. 2). The largest collinear fragment was 39,412 bp, with more than half of the fragments were greater than 5,000 bp in length. Additionally, 66 fragments were detected between R. vialii and V. macrocarpon, covering 305,580 bp (40.2%) and 257,541 bp (56%) of their respective mitogenomes. Moreover, 52 collinear fragments were identified between R. nivale subsp. boreale and V. macrocarpon, spanning 258,763 bp (50.7%) and 260,243 bp (56.6%) of their respective mitogenomes (Fig. 5). Notably, the mitogenome structure of Rhododendron was highly variability, as evidenced by the extensive rearrangements in the mitogenomes of R. nivale subsp. boreale and R. vialii.
Fig. 5.
Collinearity of the mitogenome among R. nivale subsp. boreale, V. macrocarpon, and R. vialii. Bars indicate the mitogenomes, and the light gray ribbons indicate homologous fragments (> 300 bp)
Phylogenetic analysis
We assembled and annotated the organelle genomes of two Rhododendron species, which inhabit significantly different environments. R. nivale subsp. boreale occupied mountaintop regions above an altitude of 4,000 m, while R. vialii was distributed in areas below 1,800 m. To investigate the potential role of organelle genomes in plant adaptation to different altitudes, we collected a total of 58 angiosperm organelle genomes from various altitudes, belonging to 11 families. We first explored the phylogenetic relationships based on these genomes.
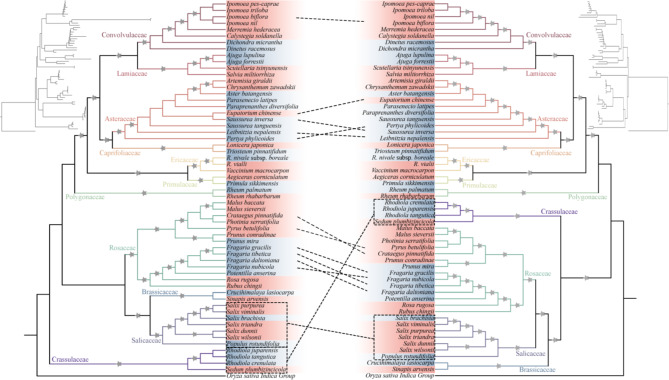
The best nucleotide substitution model was GTR + F + R3 for plastome alignment and GTR + F + I + R4 for mitogenome alignment. We constructed ML phylogenetic trees based on the PCGs of plastome and mitogenome, respectively (Fig. 6). In the mitogenome-based results, all the 11 families were monophyletic with the high support values (bootstrap = 100), and most nodes exhibited strong support (Fig. S12). Low support values were found in groups that diversified rapidly over a short period, such as Asteraceae. Compared to the mitogenome, the phylogenetic relationships in the plastome were more robust, with bootstrap values greater than 90 for almost all nodes (Fig. S13). This increased robustness may result from the larger number of genes in the plastome, providing a more comprehensive and diverse evolutionary dataset. In the plastome-based phylogeny, three nodes with low support (< 60) were found in Rosaceae and Asteraceae. Both analyses consistently showed that Rhododendron and Vaccinium were sister with 100 bootstrap values and were clustered with Primulaceae.
Fig. 6.
Consistency and conflict between mitogenome-based (left) and plastome-based (right) phylogenetic relationships. The Maximum Likelihood (ML) trees were constructed based on concatenation-based analysis of PCGs, including 58 eudicots and 1 outgroup (Oryza sativa). The five-pointed star on the branch represents the bootstrap value. The figures in the top left and right represent the phylogram of the same tree showing branch lengths. The highlighted blocks represent the altitudinal distribution of the species. Blue blocks represent the high-altitude distribution and orange blocks represent the low altitude distribution. The gray stars indicate bootstrap values greater than 95
The topological structure of the mitogenome- and plastome-based phylogenetic trees are expected to be consistent (Fig. 6). However, our results revealed obvious conflicts between the two analyses. At the family level, the main conflicts occurred among Rosaceae, Salicaceae, Brassicaceae, and Crassulaceae. In the mitochondria-based results, Crassulaceae was identified as a sister group to the other 10 families. In the chloroplast-based results, Crassulaceae was clustered with Rosaceae, Salicaceae, Brassicaceae, forming a sister group to these families. Moreover, the phylogenetic position of Salicaceae differed between the two trees: in the mitogenome-based tree, Salicaceae was the sister of Brassicaceae, whereas in the plastome-based tree, it was the sister of Rosaceae. Phylogenetic conflicts at the genus or species level primarily occurred in Asteraceae and Rosaceae. Notably, Saussurea was not monophyletic.
PCGs codon usage analysis
We analyzed the relative synonymous codon usage (RSCU) values of all PCGs of R. nivale subsp. boreale and R. vialii to evaluate the codon usage bias in their organelle genomes (Fig. S14). The range of RSCU values in the mitogenome varied from 0.44 (GCU) to 1.81 (CUU), whereas in the plastome, the range was from 0.32 (CGC) to 2.18 (UUA). The RSCU values of AUG (start codon) and UGG were equal to one in all organelle genomes. Compared to R. nivale subsp. boreale, the organelle genomes of R. vialii exhibited slightly more codon usage bias (RSCU value > 1) (R. nivale subsp. boreale: R. vialii, mitogenome = 28 : 29, plastome = 30 : 31). Additionally, no significant differences were observed in the RSCU values of the plastomes between the two species. However, several codons in the mitogenome, such as AGG, CUU, and UAA, displayed species-specific biases.
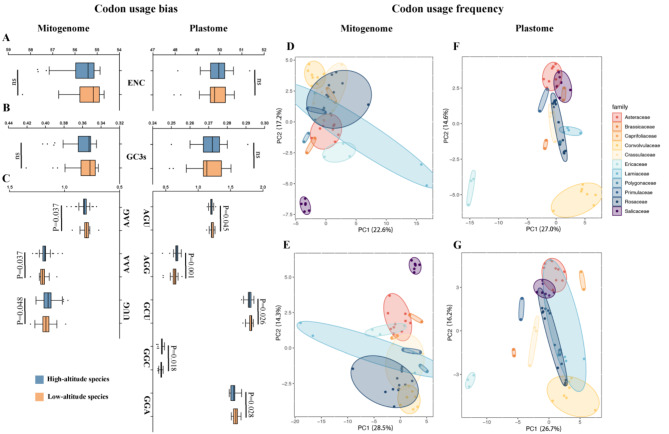
Codon usage bias is influenced by various factors, including mutational bias, selection, and GC-biased gene conversion. Therefore, habitat differences due to altitude changes may affect plant codon usage. Based on our collection of organelle genomes from species in 11 plant families, we divided them into high-altitude and low-altitude groups to explore the effect of altitude on plant codon usage bias (Fig. 6). Two important indicators, ENc and GC3s, were used to preliminarily assess the relationship between plant codon usage bias and altitude. However, there was no significant difference in ENc or GC3s between high and low altitudes (Fig. 7A, B). Subsequently, we calculated the RSCU value to detect differences in synonymous codon usage preferences at different altitudes. The results indicated no preference for most synonymous codons between different altitudes (Fig. 7C). Only a few codons were found to be related to altitude. In mitogenome, codon usage bias for UUG, AAA, and AAG differed significantly between high and low altitudes, while in the plastome, it was AGU, AGG, GCU, GGC, and GGA.
Fig. 7.
Codon usage bias in organelle genomes of 58 species. (A) The effective number of codons (ENc) distribution in high and low altitude species. (B) The GC content at the third position of codons (GC3s) distribution in high and low altitude species. (C) Relative synonymous codon usage (RSCU) values showing significant differences between high and low altitude species. (D) The PCA plot based on absolute codon usage frequency (absCUFs) of mitogenomes. (E) The PCA plot based on synonymous codon usage frequency (synCUFs) of mitogenomes. (F) The PCA plot based on absolute codon usage frequency (absCUFs) of plastomes. (G) The PCA plot based on synonymous codon usage frequency (synCUFs) of plastomes. ns represents no significance. P represents P-value from the Wilcoxon Signed-Rank Test
In addition, we evaluated the factors influencing the frequencies of codon usage using PCA. Two types of codon usage frequency, absCUFs and synCUFs, were used for analysis. All genomes were grouped based on altitude and evolutionary lineage. Several discernible patterns emerged from the analysis of these graphs: (1) When grouped based on altitude, there was no clear separation between absCUFs and synCUFs at high and low altitudes (Fig. S15); (2) When grouped by evolutionary lineages, most samples formed visibly distinct clusters based on either their absCUFs or synCUFs, although there were substantial overlaps (Fig. 7D-G); (3) The mitogenomes were not as well separated as the plastomes; (4) The codon usage frequencies of mitogenome PCGs in Salicaceae showed significant separation from other samples; (5) For the plastome PCGs, significant separations were observed in Ericaceae and Convolvulaceae.
ENc-GC3 scatter plots were frequently used to estimate factors influencing codon usage patterns (Figs. S16–S37). We performed ENc plot analysis of organelle genomes from all species to investigate the effects of mutation and selection on codon usage patterns. We found that all genomes exhibited similar bias patterns: a minority of genes were positioned on or near the standard curve, while the majority showed deviations. This suggests that the codon usage bias of most genes was influenced by factors beyond mutations. In most species, atp9 and rps13 from the mitogenome, and psbI and psbJ from the plastome, deviated significantly further from the expected curve, indicating that they were subjected to intense selection pressure, likely related to environmental adaptation.
Selection analysis
Organelles play an important role in the survival of individuals. We tested the selective pressure on each organelle PCG using the aBSREL model with an exploratory analysis. aBSREL is an improved version of the commonly used “branch-site” models, which are designed to test if positive selection has occurred on a proportion of branches. Our results indicated that atp4, atp9, mttB, ccmFN, rpl5, and rpl16 from the mitogenome and clpP, ndhA, ndhK, petD, rps3, and rps4 from the plastome were positively selected in high-altitude species (Table S25). Notably, atp4, atp9, mttB, and clpP were positively selected in multiple high-altitude species, suggesting they may play a significant role in high-altitude adaptation of plants and represent molecular convergent adaptation strategies. These genes are mainly involved in ATP production, proton transmembrane transfer, protein synthesis, and degradation of misfolded proteins.
Discussion
Plant organelles, including mitochondria and chloroplasts, are essential sites for respiration and photosynthesis, driving the accumulation of biomass [1]. Therefore, studying organelle genomes is crucial for understanding plant heredity, evolution, and adaptation. In this study, we evaluated the effectiveness of six mainstream organelle genome assembly approaches on complex genomes using diverse data sets from Rhododendron species. The results indicate that the use of HiFi data for organelle genome assembly is an efficient method. Compared to strategies using only NGS data or a combination of NGS and ONT data, HiFi reads facilitate faster and more successful assembly of organelle genomes.
In organelle genomes, the complex conformation of the mitogenome makes it more difficult to assemble than plastomes [3]. Through various tests, two complete mitogenomes of Rhododendron were eventually obtained, specifically R. nivale subsp. boreale and R. vialii. The reads mapping shows that our assembly results are gap-free, high-quality. The mitogenome of R. vialii exhibits a typical circular structure, while R. nivale subsp. boreale consists of two circular chromosomes and one linear chromosome. Compared with the previous two Rhododendron species linear mitochondrial scaffolds (R. × pulchrum: 816,410 bp; R. simsii: 802,707 bp) constructed using short reads, our assemblies are smaller [14, 15]. This size difference may result from the shorter read lengths of NGS, which can lead to redundancy in repetitive and complex regions, producing larger assemblies and complicating the retrieval of complete genomes. In addition, the similar numbers of PCGs, rRNAs and tRNAs also indicate the reliability of our assembly. Conformational diversity and intricate genomic rearrangements occur significantly in plant mitogenomes [51]. Based on long-reads mapping validation and collinearity analysis, we have confirmed the high diversity of mitochondrial genomes in species of the Ericaceae family, potentially driven by the presence of long repeat sequences. Such mitogenomic reconfigurations are frequently observed in closely related plant species, even among different varieties [52, 53].
Plastomes are relatively small and highly conserved [5]. In this study, we provide the plastomes for R. nivale subsp. boreale and R. vialii. Like most plastomes, they exhibit a tetrad structure [54]. Notably, the plastome of Rhododendron has IR regions that are twice the size of those in most other angiosperms, resulting in a larger genome size [55]. The presence of extremely long repetitive sequences in Rhododendron may be a key factor contributing to the difficulty in assembling the plastome [56]. By using HiFi data, which spans repetitive regions and ensures sequence accuracy, we successfully obtained the complete plastomes of two Rhododendron species. Although our two assemblies have similar gene numbers, the boundaries of the IRs in R. vialii have shifted, reducing the size of the SSC to only 73 bp, a phenomenon also observed in the plastomes of Corydalis and Paphiopedilum [57, 58]. These large alterations are associated with DNA double-strand breaks and subsequent repair processes, likely resulting in variations in genome size, gene count, and the emergence of pseudogenes, reflecting the evolutionary processes of plants [59].
The mitogenome is frequently inserted with sequences from the plastome, typically constituting 1–12% of the total mitochondrial size [60]. In our study, plastome transfer to mitogenome was more pronounced in R. vialii, comprising 3.26% of its mitochondrial genome, compared to 2.37% in R. nivale subsp. boreale. These horizontal gene transfers facilitate gene flow between organelles and act as a significant driving force in evolution [61]. Although the proportion is small, it still includes several function genes. However, most organelle gene transfers are nonfunctional [62]. Whether these genes perform specific functions in the mitochondria requires further verification.
In angiosperms, the nuclear genome, plastome, and mitogenome constitute three distinct sets of hereditary material [63]. Nonetheless, the structure of phylogenetic trees derived from these genomes is inconsistent, fluctuating with the choice of species, data sets, and the extent of the data [64]. To investigate the role of organelles in plant adaptation to high altitudes, organelle genomes from 58 species across 11 families, representing different altitudes, were collected. Phylogenetic relationships were reconstructed using the ML method, and the results indicated strong support for most clades, with all families being monophyletic. The clade comprising R. nivale subsp. boreale, R. vialii, and V. macrocarpon is identified as part of the Ericaceae, supported by high support. The Primulaceae was identified as a sister clade to the Ericaceae. This relationship has been verified through phylogenetic analyses using two separate organelle genomes and is consistent with previous findings [14]. However, phylogenetic structures derived from various genomic datasets exhibit discrepancies, such as Crassulaceae and Salicaceae, reflecting existing phylogenetic patten [65]. These topological inconsistencies between independently inherited genomic datasets may be attributed to differential species sampling and incomplete evolutionary lineage sorting [66, 67]. In addition, differences in the inferred evolutionary model and hybridization events are also considered contributing factors [68, 69]. Relationships within families are more complex, especially in some species-rich groups with a short period of radiation evolution, such as Asteraceae. To further resolve the complex relationships between these species, larger and higher-resolution datasets are needed [70].
Further, we used these genomes to speculate on possible high-altitude adaptation strategies in angiosperms. Codon usage bias, which refers to the preferential use of specific nucleotide triplets (codons) to encode amino acids in the PCGs of a species, is influenced by mutational pressure, natural selection, and GC-biased gene conversion [71]. Organisms from similar environmental background may exhibit common codon usage preferences due to their shared evolutionary history. Therefore, exploring codon usage patterns is instrumental in understanding the processes of environmental adaptation and the intricate molecular mechanisms contributing to the rich diversity of life [72]. Our study found no significant differences in codon usage bias (ENc and GC3s) among species at different altitudes, suggesting a weak influence of altitude on codon usage in angiosperms. To explore this subtle effect, we compared the RSCU values between high- and low-altitude species. The majority of RSCU values showed no differences between altitudes, with only 8 exceptions. Although rare, these variations may play a pivotal role in facilitating the adaptive success of species within their respective environments through the modulation of gene expression levels [73].
Codon usage frequencies are considered to potentially reveal evolutionary mechanisms underlying biological adaptation to various environments [45]. However, PCA indicates that codon usage frequency patterns are primarily associated with phylogenetic lineages, which probably significantly surpass environmental factors. Consistent with our hypothesis, the impact of the environment on codon usage appears to be limited. ENc-plot analysis further supports this view. Similar codon usage patterns are more frequently observed among closely related species, with consistent features evident in other closely related species such as Elaeagnus and Aconitum [74, 75]. Although altitude variation represents an important example of environmental differences, additional evidence from diverse environments is necessary. Furthermore, the divergence in codon usage frequencies among different evolutionary lineages within the plastome is more pronounced, likely associated with the heterogeneous rates of evolution between the plastome and mitogenome [76]. Overall, in this study, codon usage is not strongly associated with plant survival at high and low altitudes. Phylogenetic factors likely play a key role in determining the fundamental patterns of codon usage, while environmental factors further fine-tune and optimize these patterns, like the rare RSCU variations. Recent studies have also showed that natural selection is only a secondary factor at the synonymous substitution [77].
Mitochondria and chloroplasts are crucial for life processes, and detecting their molecular adaptation strategies enhances our understanding of evolution [78]. Using the aBSREL model, we examined gene selection in mitogenome and plastome across 58 species from various altitudes to hypothesize about high-altitude adaptation strategies of organelle genes. Four candidate genes for high-altitude adaptation were identified: three (atp4, atp9, mttB) from the mitogenome and one (clpP) from the plastome. The genes atp4 and atp9 are key components of ATP synthase synthesis [79, 80]. In addition, atp4 may be involved in plant growth and development, while the atp9 directly affects pollen development [81, 82]. High-altitude environments are harsh, with pollinators often scarce [83].The positive selection of atp4 and atp9 likely facilitates rapid ATP synthesis, improves plant growth and development, enhances pollen viability, thereby ensuring survival in extreme conditions. The mttB gene encodes a membrane transport protein [84]. Although its function is not entirely clear, it is considered indispensable, as it is rarely lost in angiosperms [85]. Additionally, in other high-altitude plants like Elymus sibiricus, numerous transcriptional elements have been identified around mttB, which are likely induced by environmental stress related to plateau adaptation, suggesting that mttB may be related to high-altitude adaptation [86]. The clpP gene is crucial for protease synthesis and plays a major role in degrading misfolded proteins [87]. Altitude correlates positively with UV radiation, increasing by 5.1–15% every 1,000 m [88]. Intense UV radiation generates reactive oxygen species, damaging biological macromolecules and altering protein conformation and function [89, 90]. Positive selection on the clpP gene likely facilitates efficient removal of conformationally altered proteins induced by adverse environmental conditions in high-altitude plants. Therefore, atp4, atp9, mttB, and clpP can be considered candidate genes for organelle adaptation to high altitudes, warranting further verification.
Conclusion
In this study, we tested various mitogenome and plastome assembly approaches using different datasets and successfully assembled and annotated organelle genomes of R. nivale subsp. boreale and R. vialii. HiFi reads were used to verify the different conformations of the mitogenome. Based on our assemblies, feature analysis was performed, including RNA editing, repeat sequences, plastome and mitogenome homologous segments, and collinearity analysis. Furthermore, after collecting organelle genomes of 58 species from different altitudes, we explored the consistency and conflict in their phylogenetic relationships and investigated the potential molecular mechanisms of high-altitude adaptation of organelles, focusing on PCGs codon usage and gene positive selection. Our results indicate that two families and several species exhibit conflict between mitogenome and plastome-based phylogenetic outcomes. Moreover, codon usage is more strongly correlated with evolutionary lineage, while the environment may act as a weak regulatory factor. Four genes (atp4, atp9, mttB, and clpP) have been positively selected in multiple high-altitude species, likely playing important roles in high-altitude adaptation. Overall, this study evaluated methods suitable for the assembly of complex organelle genomes and provides insights into the role of organelles in the molecular mechanisms underlying high-altitude adaptation in angiosperms.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank all the project members for their contributions and BGI for providing the sequencing services.
Author contributions
Z.Y. Lyu: wrote the manuscript and data analysis. S.K. Shen: reviewed and revised the manuscript. G.M. Yang: collected data. S.Q. Wang: collected samples. X.L. Zhou: collected samples. R. Zhang: collected samples.
Funding
This study was supported by the National Natural Science Foundation of China (32360099, 32471600), the Science and Technology Fund of Yunnan Province (202304BI090005), the funding of Yunnan International Joint Laboratory(R&D Center)of plateau woody vegetables development and utilization (202403AP140010), the Key Industry Science and Technology Service Project of Higher Education of Yunnan Province (FWCY-BSPY2024031) and the Scientific Research Fund of Yunnan Provincial Education Department (2024Y003).
Data availability
The raw sequencing data of this study have been deposited in the Sequence Read Archive (SRA) under Bioproject number PRJNA1040959. The genome assemblies, annotation data, and internal scripts are available at Figshare (https://doi.org/10.6084/m9.figshare.25909855). The genomes have also been deposited in NCBI GenBank with accession numbers PQ537102, PQ537103, PQ537104, PQ537097, PQ536991 and PQ536992, respectively.
Declarations
Ethics approval and consent to participate
Permissions for the collection of the plant specimens were obtained from the relevant authorities. The plant material used in this study adhered to relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang J, Kan S, Liao X, Zhou J, Tembrock LR, Daniell H, Jin S, Wu Z. Plant organellar genomes: much done, much more to do. Trends Plant Sci. 2024;2024:1360–85. [DOI] [PubMed] [Google Scholar]
- 2.Xu JP. The inheritance of organelle genes and genomes: patterns and mechanisms. Genome. 2005;48(6):951–8. [DOI] [PubMed] [Google Scholar]
- 3.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 2015;112(33):10177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozik A, Rowan BA, Lavelle D, Berke L, Schranz ME, Michelmore RW, Christensen AC. The alternative reality of plant mitochondrial DNA: one ring does not rule them all. Plos Genet. 2019;15(8):e1008373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. Bmc Biol. 2009;7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Kuang TH, Dong WL, Qian ZH, Zhang HJ, Landis JB, Feng T, Li LJ, Sun YX, Huang JL, et al. Genomic convergence underlying high-altitude adaptation in alpine plants. J Integr Plant Biol. 2023;65(7):1620–35. [DOI] [PubMed] [Google Scholar]
- 8.Venzhik Y, Deryabin A, Moshkov I. Adaptive strategy of plant cells during chilling: aspect of ultrastructural reorganization. Plant Sci. 2023;332:111722. [DOI] [PubMed] [Google Scholar]
- 9.Bock DG, Andrew RL, Rieseberg LH. On the adaptive value of cytoplasmic genomes in plants. Mol Ecol. 2014;23(20):4899–911. [DOI] [PubMed] [Google Scholar]
- 10.Budar F, Roux F. The role of organelle genomes in plant adaptation: time to get to work! Plant Signal Behav. 2011;6(5):635–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YS, Deng T, Zhou Z, Sun H. Is the east Asian flora ancient or not? Natl Sci Rev. 2018;5(6):920–32. [Google Scholar]
- 12.Fu CN, Mo ZQ, Yang JB, Cai J, Ye LJ, Zou JY, Qin HT, Zheng W, Hollingsworth PM, Li DZ, Gao LM. Testing genome skimming for species discrimination in the large and taxonomically difficult genus Rhododendron. Mol Ecol Resour. 2022;22(1):404–14. [DOI] [PubMed] [Google Scholar]
- 13.Mo ZQ, Fu CN, Zhu MS, Milne RI, Yang JB, Cai J, Qin HT, Zheng W, Hollingsworth PM, Li DZ, Gao LM. Resolution, conflict and rate shifts: insights from a densely sampled plastome phylogeny for Rhododendron (Ericaceae). Ann Bot. 2022;130(5):687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J, Li X, Li M, Cheng H, Huang X, Jin S. Characterization, comparative phylogenetic, and gene transfer analyses of organelle genomes of Rhododendron x Pulchrum. Front Plant Sci. 2022;13:969765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Luo H, Nie S, Zhang RG, Mao JF. The complete mitochondrial and plastid genomes of Rhododendron Simsii, an important parent of widely cultivated azaleas. Mitochondrial DNA B Resour. 2021;6(3):1197–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radzvilavicius AL, Johnston IG. Organelle bottlenecks facilitate evolvability by traversing heteroplasmic fitness valleys. Front Genet. 2022;13:974472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie S, Zhao S-W, Shi T-L, Zhao W, Zhang R-G, Tian X-C, Guo J-F, Yan X-M, Bao Y-T, Li Z-C et al. Gapless genome assembly of azalea and multi-omics investigation into divergence between two species with distinct flower color. Hortic Res-England 2022, 10(1). [DOI] [PMC free article] [PubMed]
- 18.Chang Y, Zhang R, Ma Y, Sun W. A haplotype-resolved genome assembly of Rhododendron Vialii based on PacBio HiFi reads and Hi-C data. Sci Data. 2023;10(1):451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–6. [DOI] [PubMed] [Google Scholar]
- 22.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. Plos Comput Biol. 2017;13(6):e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi C, Shen F, Han F, Qu Y, Hou J, Xu K, Xu LA, He W, Wu Z, Yin T. PMAT: an efficient plant mitogenome assembly toolkit using low-coverage HiFi sequencing data. Hortic Res. 2024;11(3):uhae023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou W, Armijos CE, Lee C, Lu R, Wang J, Ruhlman TA, Jansen RK, Jones AM, Jones CD. Plastid Genome Assembly using long-read data. Mol Ecol Resour. 2023;23(6):1442–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(W1):W6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn NA, Unni DR, Diesh C, Munoz-Torres M, Harris NL, Yao E, Rasche H, Holmes IH, Elsik CG, Lewis SE. Apollo: democratizing genome annotation. Plos Comput Biol. 2019;15(2):e1006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Li J, Ma Y, Kou L, Wei J, Wang W. The complete mitochondrial genome of okra (Abelmoschus esculentus): using nanopore long reads to investigate gene transfer from chloroplast genomes and rearrangements of mitochondrial DNA molecules. BMC Genomics. 2022;23(1):481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H. New strategies to improve minimap2 alignment accuracy. Bioinformatics. 2021;37(23):4572–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edera AA, Small I, Milone DH, Sanchez-Puerta MV. Deepred-Mt: deep representation learning for predicting C-to-U RNA editing in plant mitochondria. Comput Biol Med. 2021;136:104682. [DOI] [PubMed] [Google Scholar]
- 34.Beier S, Thiel T, Munch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He W, Yang J, Jing Y, Xu L, Yu K, Fang X. NGenomeSyn: an easy-to-use and flexible tool for publication-ready visualization of syntenic relationships across multiple genomes. Bioinformatics. 2023;39(3):btad121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 2020;20(1):348–55. [DOI] [PubMed] [Google Scholar]
- 40.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letunic I, Bork P. Interactive tree of life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res 2024:gkae268. [DOI] [PMC free article] [PubMed]
- 44.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software suite. Trends Genet. 2000;16(6):276–7. [DOI] [PubMed] [Google Scholar]
- 45.Panda A, Tuller T. Determinants of associations between codon and amino acid usage patterns of microbial communities and the environment inferred based on a cross-biome metagenomic analysis. Npj Biofilms Microbiomes. 2023;9(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villanueva RAM, Chen ZJ. ggplot2: Elegant Graphics for Data Analysis, 2nd edition. Measurement-Interdisciplinary Research and Perspectives 2019, 17(3):160–167.
- 47.Grunsky EC. R: a data analysis and statistical programming environment - an emerging tool for the geosciences. Comput Geosci. 2002;28(10):1219–22. [Google Scholar]
- 48.Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. MACSE v2: Toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 2018;35(10):2582–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–9. [DOI] [PubMed] [Google Scholar]
- 50.Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Pond SLK. Less is more: an adaptive branch-site Random effects Model for efficient detection of episodic diversifying selection. Mol Biol Evol. 2015;32(5):1342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Hu JY, Li DZ. Remarkable mitochondrial genome heterogeneity in Meniocus Linifolius (Brassicaceae). Plant Cell Rep. 2024;43(2):36. [DOI] [PubMed] [Google Scholar]
- 52.He X, Zhang X, Deng Y, Yang R, Yu LX, Jia S, Zhang T. Structural reorganization in Two Alfalfa Mitochondrial Genome Assemblies and mitochondrial evolution in Medicago Species. Int J Mol Sci. 2023;24(24):17334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu D, Zhang Z, Hao Y, Li M, Yu H, Zhang X, Mi H, Cheng L, Zhao Y. Decoding the complete organelle genomic architecture of Stewartia gemmata: an early-diverging species in Theaceae. BMC Genomics. 2024;25(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daniell H, Jin S, Zhu XG, Gitzendanner MA, Soltis DE, Soltis PS. Green giant-a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol J. 2021;19(3):430–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 2016, 17. [DOI] [PMC free article] [PubMed]
- 56.Mascher M, Wicker T, Jenkins J, Plott C, Lux T, Koh CS, Ens J, Gundlach H, Boston LB, Tulpova Z, et al. Long-read sequence assembly: a technical evaluation in barley. Plant Cell. 2021;33(6):1888–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo YY, Yang JX, Bai MZ, Zhang GQ, Liu ZJ. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. Bmc Plant Biol. 2021;21(1):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park S, An B, Park S. Dynamic changes in the plastid and mitochondrial genomes of the angiosperm Corydalis pauciovulata (Papaveraceae). Bmc Plant Biol. 2024;24(1):303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffmann M, Kuhn J, Daschner K, Binder S. The RNA world of plant mitochondria. Prog Nucleic Acid Res Mol Biol. 2001;70:119–54. [DOI] [PubMed] [Google Scholar]
- 61.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424(6945):197–201. [DOI] [PubMed] [Google Scholar]
- 62.Cusimano N, Wicke S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2016;210(2):680–93. [DOI] [PubMed] [Google Scholar]
- 63.Qiu YL, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature. 1999;402(6760):404–7. [DOI] [PubMed] [Google Scholar]
- 64.Hu H, Sun P, Yang Y, Ma J, Liu J. Genome-scale angiosperm phylogenies based on nuclear, plastome, and mitochondrial datasets. J Integr Plant Biol. 2023;65(6):1479–89. [DOI] [PubMed] [Google Scholar]
- 65.Hu YB, Wang XP, Xu YC, Yang H, Tong ZY, Tian R, Xu SH, Yu L, Guo YL, Shi P, et al. Molecular mechanisms of adaptive evolution in wild animals and plants. Sci China Life Sci. 2023;66(3):453–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma J, Sun P, Wang D, Wang Z, Yang J, Li Y, Mu W, Xu R, Wu Y, Dong C, et al. The Chloranthus sessilifolius genome provides insight into early diversification of angiosperms. Nat Commun. 2021;12(1):6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 2014;5:4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyszka AS, Bretz EC, Robertson HM, Woodcock-Girard MD, Ramanauskas K, Larson DA, Stull GW, Walker JF. Characterizing conflict and congruence of molecular evolution across organellar genome sequences for phylogenetics in land plants. Front Plant Sci. 2023;14:1125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xue T-T, Janssens SB, Liu B-B, Yu S-X. Phylogenomic conflict analyses of the plastid and mitochondrial genomes via deep genome skimming highlight their independent evolutionary histories: a case study in the cinquefoil genus Potentilla Sensu lato (Potentilleae, Rosaceae). Mol Phylogenet Evol. 2024;190:107956. [DOI] [PubMed] [Google Scholar]
- 70.Simon C. An evolving view of phylogenetic support. Syst Biol. 2022;71(4):921–8. [DOI] [PubMed] [Google Scholar]
- 71.Parvathy ST, Udayasuriyan V, Bhadana V. Codon usage bias. Mol Biol Rep. 2022;49(1):539–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mazumdar P, Binti Othman R, Mebus K, Ramakrishnan N, Ann Harikrishna J. Codon usage and codon pair patterns in non-grass monocot genomes. Ann Bot. 2017;120(6):893–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li N, Li YY, Zheng CC, Huang JG, Zhang SZ. Genome-wide comparative analysis of the codon usage patterns in plants. Genes Genomics. 2016;38(8):723–31. [Google Scholar]
- 74.Li C, Zhou L, Nie J, Wu S, Li W, Liu Y, Liu Y. Codon usage bias and genetic diversity in chloroplast genomes of Elaeagnus species (myrtiflorae: Elaeagnaceae). Physiol Mol Biol Plants. 2023;29(2):239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang M, Liu J, Yang W, Li Z, Hai Y, Duan B, Zhang H, Yang X, Xia C. Analysis of codon usage patterns in 48 Aconitum species. BMC Genomics. 2023;24(1):703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. P Natl Acad Sci USA. 1987;84(24):9054–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clement Y, Sarah G, Holtz Y, Homa F, Pointet S, Contreras S, Nabholz B, Sabot F, Saune L, Ardisson M, et al. Evolutionary forces affecting synonymous variations in plant genomes. Plos Genet. 2017;13(5):e1006799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alvarez-Carretero S, Kapli P, Yang Z. Beginner’s guide on the Use of PAML to detect positive selection. Mol Biol Evol. 2023;40(4):msad041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heazlewood JL, Whelan J, Millar AH. The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 2003;540(1–3):201–5. [DOI] [PubMed] [Google Scholar]
- 80.Meyer EH, Taylor NL, Millar AH. Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J Proteome Res. 2008;7(2):786–94. [DOI] [PubMed] [Google Scholar]
- 81.Reddemann A, Horn R. Recombination events involving the atp9 gene are Associated with male sterility of CMS PET2 in sunflower. Int J Mol Sci. 2018;19(3):806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang G, Zhong MY, Shuai BL, Song JD, Zhang J, Han L, Ling HL, Tang YP, Wang GF, Song RT. E plus subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 2017;214(4):1563–78. [DOI] [PubMed] [Google Scholar]
- 83.Shen SK, Zhou XL, Wang SQ, Lyu ZY, Zhang R, Liu Y, Long B. Protect fragile mountaintop ecosystems. Science. 2023;380(6650):1114–5. [DOI] [PubMed] [Google Scholar]
- 84.Xu L, Wang J, Zhang T, Xiao H, Wang H. Characterizing complete mitochondrial genome of Aquilegia amurensis and its evolutionary implications. Bmc Plant Biol. 2024;24(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 2002;99(15):9905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiong YL, Yu QQ, Xiong Y, Zhao JM, Lei X, Liu L, Liu W, Peng Y, Zhang JB, Li DX, et al. The complete mitogenome of Elymus sibiricus and insights into its evolutionary pattern based on simple repeat sequences of seed plant mitogenomes. Front Plant Sci. 2022;12:802321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sjögren LLE, Stanne TM, Zheng B, Sutinen S, Clarke AK. Structural and functional insights into the chloroplast ATP-dependent clp protease in Arabidopsis. Plant Cell. 2006;18(10):2635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blumthaler M, Ambach W, Ellinger R. Increase in solar UV radiation with altitude. J Photoch Photobio B. 1997;39(2):130–4. [Google Scholar]
- 89.Akter S, Huang J, Waszczak C, Jacques S, Gevaert K, Van Breusegem F, Messens J. Cysteines under ROS attack in plants: a proteomics view. J Exp Bot. 2015;66(10):2935–44. [DOI] [PubMed] [Google Scholar]
- 90.Fraikin GY. Signaling mechanisms regulating diverse plant cell responses to UVB Radiation. Biochem (Mosc). 2018;83(7):787–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data of this study have been deposited in the Sequence Read Archive (SRA) under Bioproject number PRJNA1040959. The genome assemblies, annotation data, and internal scripts are available at Figshare (https://doi.org/10.6084/m9.figshare.25909855). The genomes have also been deposited in NCBI GenBank with accession numbers PQ537102, PQ537103, PQ537104, PQ537097, PQ536991 and PQ536992, respectively.