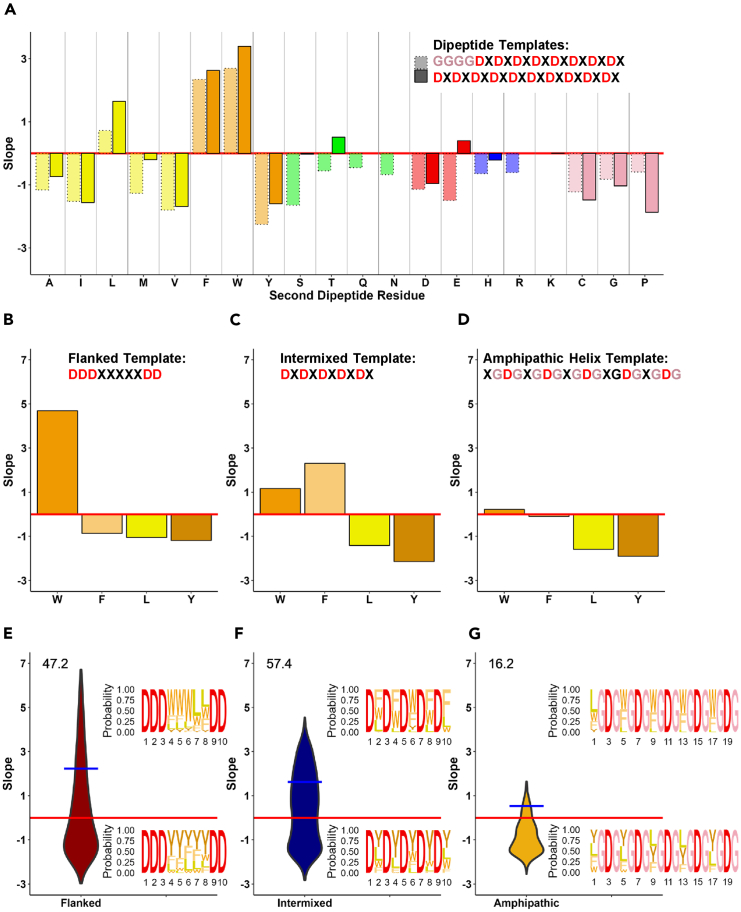
Figure 2.
Functionality of balanced acidic/hydrophobic sequences in different arrangement contexts
(A) Functionality of dipeptide repeats with an aspartic acidic (D) residue in the first position of the dipeptide. Two dipeptide templates: 8 repeats (dotted borders) and 10 repeats (solid borders). X axis: individual amino acid used in X positions (blue – basic, red – acidic, green – hydrophilic neutral, yellow/orange hydrophobic, pink – others); Y axis: growth slope of cells carrying the individual sequences.
(B–G) Functionality of sequences with only one type of hydrophobic residue (W, F, L, or Y) within three sequence templates: flanked template (DDDXXXXXDD, panel B), intermixed template (DXDXDXDXDX, panel C), and amphipathic helix template (XGDGXGDGXGDGXGDGXGDG, panel D). Axes same as in A. (E–G) Distributions of AD activities of mixed amino acids (W, F, L, and Y) within five X positions in each sequence template: flanked template (panel E), intermixed template (panel F), and amphipathic helix template (panel G) (1024 individual sequences for each template). X axis: template used; Y axis: same as in A. Values above violin plots indicate percentage of functional sequences within each template dataset. Blue bar indicates the average growth slope for sequences above the 0 threshold. Inset sequence logos depict proportion of each amino acid at each position, for the top 5% (top) and bottom 5% (bottom) of sequences for each template dataset.