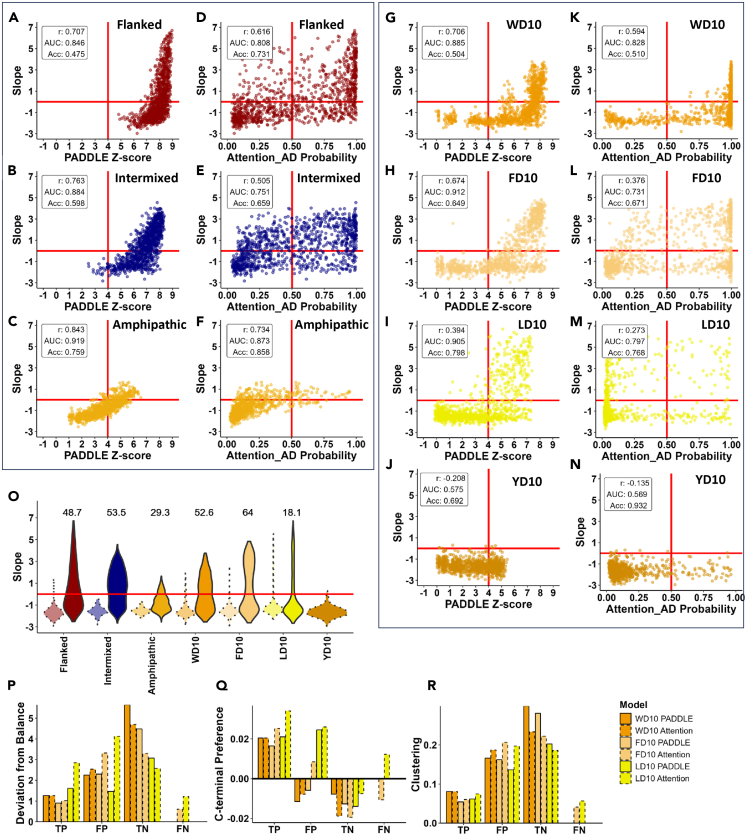
Figure 7.
Performance and comparison of ML models PADDLE and Attention AD, and Mechanistic Predictor
(A–N) PADDLE ML model7 (panels A, B, C, G, H, I, & J) and Attention AD model22 (panels D, E, F, K, L, M, & N) used to predict functionality across seven sets of sequences: Flanked WYFL (panels A-B), Intermixed WYFL (panels B-E), Amphipathic Helix WYFL (panels C–F), WD10 (panels G–K), FD10 (panels H–L), LD10 (panels I–M), and YD10 (panel J–N). X axis: Z-scores represent predicted probability of functionality using PADDLE (see STAR Methods) with scores greater than a threshold of 4 predicted to be functional.7 Probability values represent predicted probability of functionality using Attention_AD with scores greater than a threshold of 0.5 predicted to be functional. Y axis: growth slope of cells carrying the corresponding sequences. Values in the upper left corner of each panel are the correlation between the growth slope and prediction (r), area under the ROC curve (AUC) and the fraction accurately predicted (Acc).
(O) A modified Mechanistic Predictor6 used to predict functionality across seven sets of sequences. The modified predictor applied to sequences with a total length of 20 residues: Functional ADs = [-6.5 ≤ Net Charge ≤ −4 & Number of W, F, and L Residues ≥3]. Distributions of sequences predicted to be functional (solid borders) and sequences predicted to be non-functional (dotted borders). X axis: dataset used; Y axis: same as in A. Values on the graph represent the percent of experimentally determined functional sequences out of the sequences predicted to be functional using the modified mechanistic predictor.
(P–R) Average scores for three rules: balance (panel P), position (panel Q), and clustering (panel R) within sequences correctly predicted (true positives (TP) and true negatives (TN)) and incorrectly predicted (false positives (FP) and false negatives (FN)) to be activation domains by PADDLE (solid borders) and Attention AD (dotted borders). Deviation from balance is the absolute value of the difference in number of acidic and hydrophobic residues. C-terminal preference is the slope calculated from a best fit line where the X axis was 1–10 for the ten positions of each sequence and the Y axis was the number of sequences that had a hydrophobic residue at each position, with a greater preference indicating more hydrophobic residues toward the C-terminus (see Figures 4 panels H, I, J).