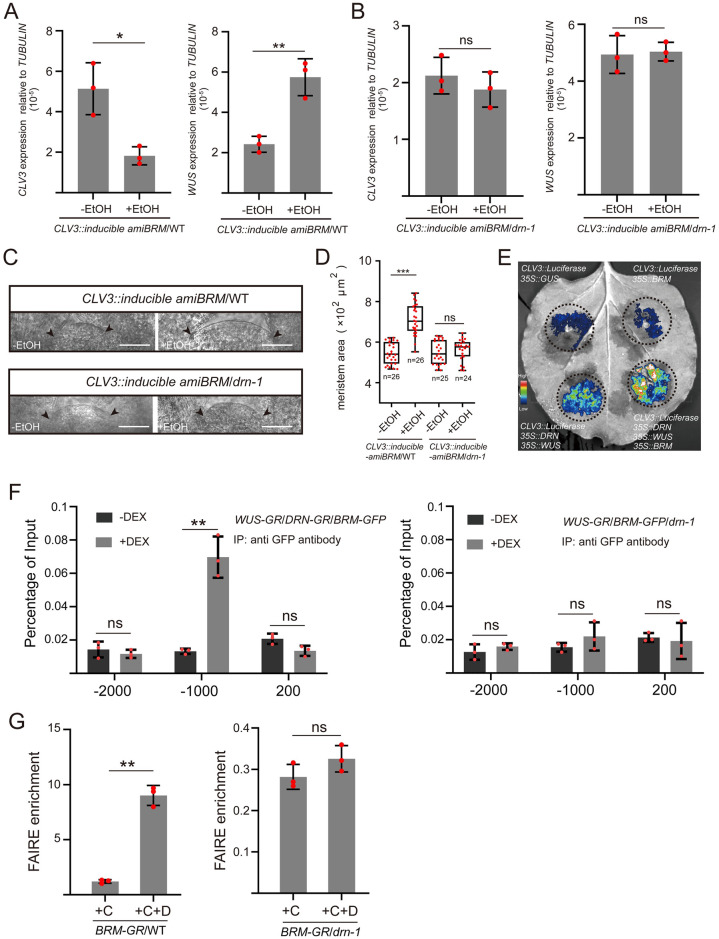
Fig 6. The positive regulation of CLV3 by BRM relies on DRN.
(A) qRT-PCR was applied to test the relative transcript levels of CLV3 and WUS after ethanol (EtOH) induction for 24 h in CLV3::inducible amiBRM/WT transgenic seedlings (14-day-old). Two independent experiments were performed with similar results. (B) qRT-PCR was applied to test the relative transcript levels of CLV3 and WUS after ethanol (EtOH) induction for 24 h in CLV3::amiBRM/drn-1 transgenic seedlings (14-day-old). Two independent experiments were performed with similar results. (C) SAMs of the mock group and EtOH treated group (72 h) in CLV3::inducible amiBRM/WT and CLV3::inducible amiBRM/drn-1 transgenic seedlings (14-day-old). Black arrows indicate the boundaries of SAMs. Bars, 50 μm. Two independent experiments were performed with similar results. (D) Areas of SAMs in C were measured by Image J software. Two independent experiments were performed with similar results. (E) Different combinations of vectors, including CLV3::Luciferase, 35S::GUS, 35S::WUS, 35S::DRN, and 35S::BRM were transformed into tobacco leaves via Agrobacterium. The intensity of luciferase signal represents the activity of CLV3 promoter. The regions where Agrobacterium transformed were circled by dotted lines. Two independent experiments were performed with similar results. (F) UBQ10::mCherry-WUS-GR/35S::DRN-GR/UBQ10::BRM-GFP lines (14-day-old seedlings) were used for ChIP assays. Upon DEX induction, the nuclear localization of WUS-GR enables the association of the CLV3 promoter (WUS-binding site, from −1,082 to −1,079) by BRM-GFP, using the anti-GFP antibody for IP, but not found in UBQ10::mCherry-WUS-GR/UBQ10::BRM-GFP/drn-1 lines. The upstream −2,000 bp site and downstream +200 site acted as negative control loci (no binding site). Two independent experiments were performed with similar results. (G) DNA accessibility at the CLV3 locus (WUS-binding site −1,080) was checked by FAIRE assays after DEX treatment, using UBQ10::BRM-GR/WT and UBQ10::BRM-GR/drn-1 lines (10-day-old seedlings). The ratio of FAIRE enrichment at the CLV3 promoter (WUS-binding site −1,080) was normalized to Ta3 retrotransposon. +C, cycloheximide; +D, dexamethasone. Two independent experiments were performed with similar results. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significant difference; Student’s t test in A, B, D, F, and G. Data represent means ± SDs from 3 biological replicates in A, B, F, and G. The data underlying this figure can be found in S1 Data. ChIP, chromatin immunoprecipitation; DEX, dexamethasone; FAIRE, formaldehyde-assisted isolation of regulatory elements; SAM, shoot apical meristem.