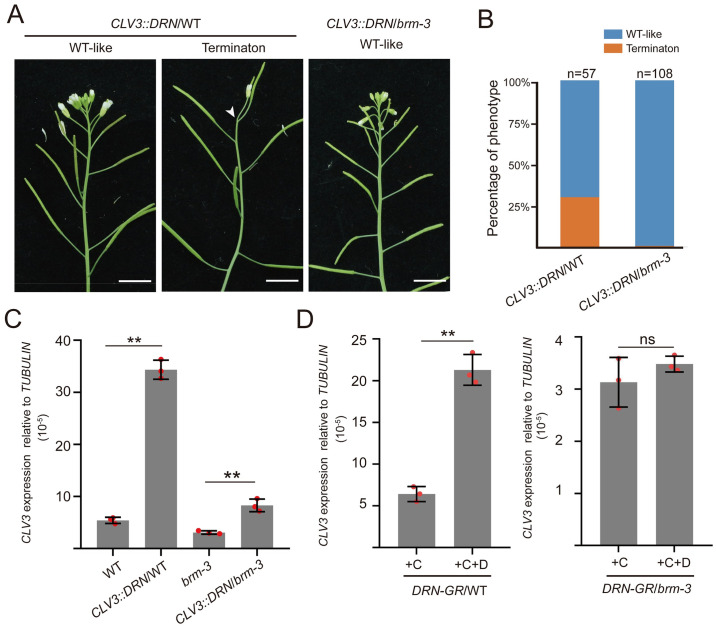
Fig 7. The activation of CLV3 expression by DRN requires BRM.
(A, B) The phenotypes of CLV3::DRN transgenic plants in WT and brm-3 backgrounds are shown and statistically analyzed. The plants in A were analyzed in B. Two independent experiments were performed with similar results. (C) qRT-PCR was used to test the relative transcript level of CLV3 in CLV3::DRN/WT, brm-3, and CLV3::DRN/brm-3 lines, using 14-day-old seedlings. Two independent experiments were performed with similar results. (D) qRT-PCR was applied to test the relative CLV3 transcript level of CLV3::DRN-GR in the wild-type background or brm-3 mutant background after DEX induction compared with the mock group, using 14-day-old seedlings. +C, cycloheximide; +D, dexamethasone. Two independent experiments were performed with similar results. **P < 0.01; ns, no significant difference; Student’s t test in C, and D. Data represent means ± SDs from three biological replicates in C and D. The data underlying this figure can be found in S1 Data. DEX, dexamethasone.