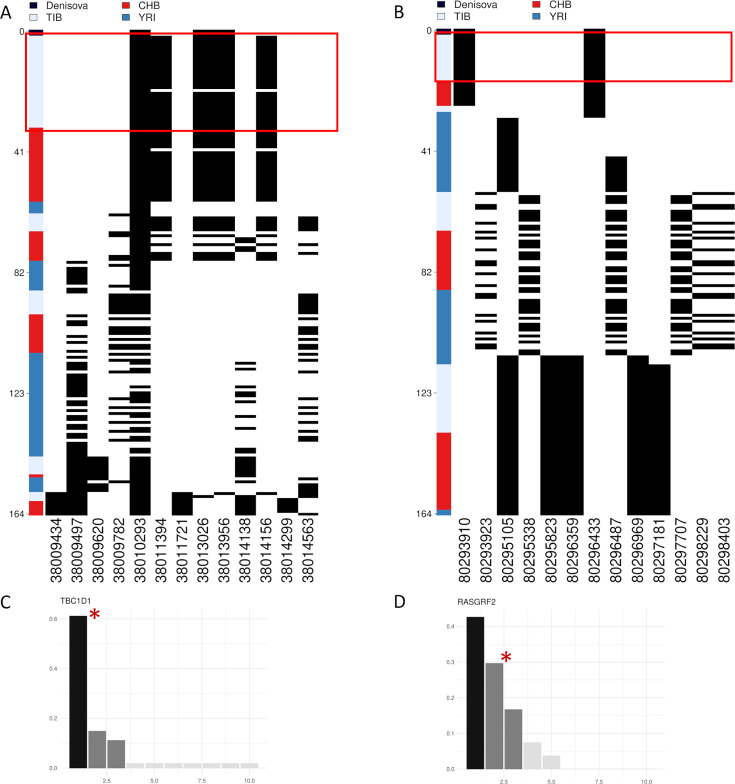
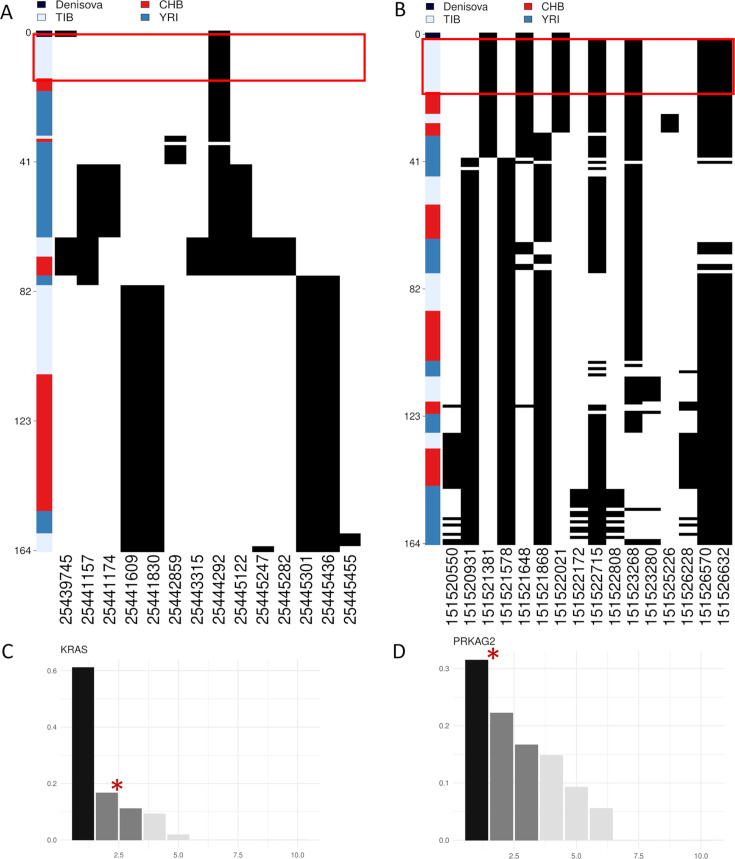
Figure 4. Representation of genetic distances between modern and archaic haplotypes and barplots showing haplotype frequency spectra for TBC1D1 and RASGRF2 candidate adaptive introgression (AI) genes.
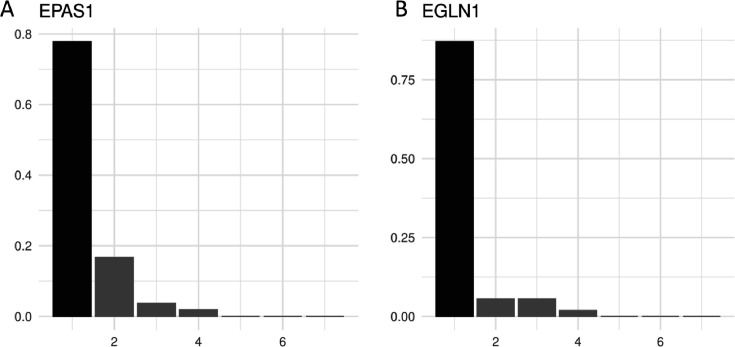
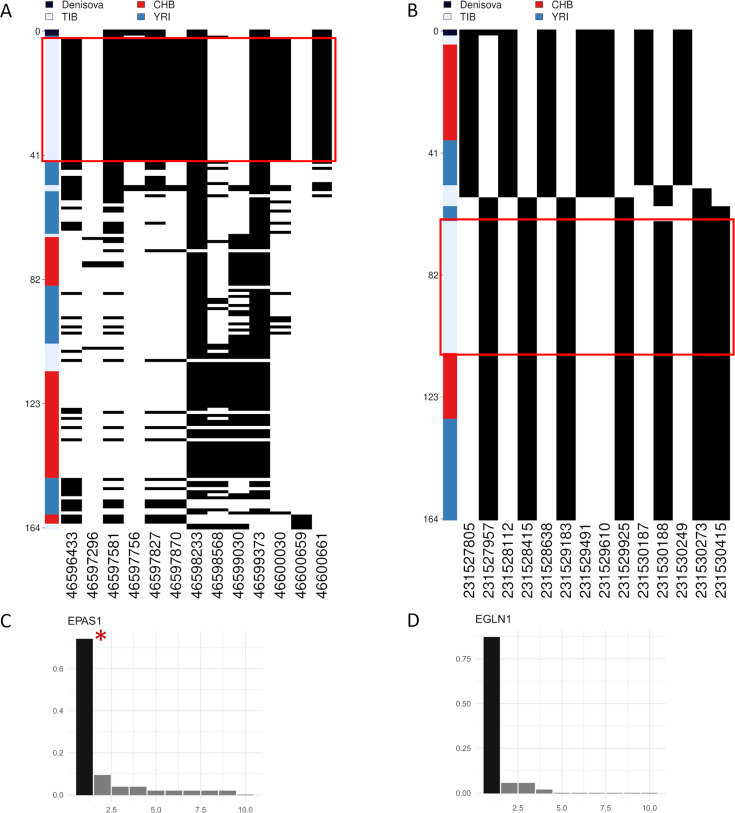
Haplotypes are reported in rows, while derived (i.e., black square) and ancestral (i.e., white square) alleles are displayed in columns. Haplotypes are ranked from top to bottom according to their number of pairwise differences with respect to the Denisovan sequence. (A) Heatmap displaying divergence between Tibetan, CHB and YRI TBC1D1 haplotypes with respect to the Denisovan genome. A total of 33 TBC1D1 haplotypes (i.e., 61% of the overall haplotypes inferred for such a region) belonging to individuals with Tibetan ancestry are plotted in the upper part of the heatmap thus presenting the smallest number of pairwise differences with respect to the Denisovan sequence. (B) Heatmap displaying divergence between Tibetan, CHB and YRI RASGRF2 haplotypes with respect to the Denisovan genome. A total of 16 Tibetan haplotypes in the RASGRF2 genomic region present no differences with respect to the Denisovan sequence. As regards barplots, on the x-axis are reported the haplotypes detected in the considered genomic windows, while on the y-axis is indicated the frequency for each haplotype. The black and dark-grey bars indicate the more frequent haplotypes (i.e., the putative adaptive haplotypes inferred by the LASSI method), while red stars mark those haplotypes carrying Denisovan-like derived alleles. (C) TBC1D1 haplotype frequency spectrum. The TBC1D1 gene presents a haplotype pattern qualitatively comparable to that observed at EPAS1 (Figure 4—figure supplement 3A), with a predominant haplotype carrying archaic derived alleles and reaching elevated frequencies in Tibetan populations. In line with this observation, such a pattern was inferred by LASSI as conformed with a non-neutral evolutionary scenario, even if it seems to be characterized by a soft rather than a hard selective sweep due to the occurrence of three sweeping haplotypes. (D) RASGRF2 haplotype frequency spectrum. A soft selective sweep was inferred also for the considered RASGRF2 genomic window, although frequencies reached by the sweeping haplotypes turned out to be more similar with each other. The second most represented haplotype was that carrying the archaic derived alleles and, reached a frequency of 29% in the Tibetan group.