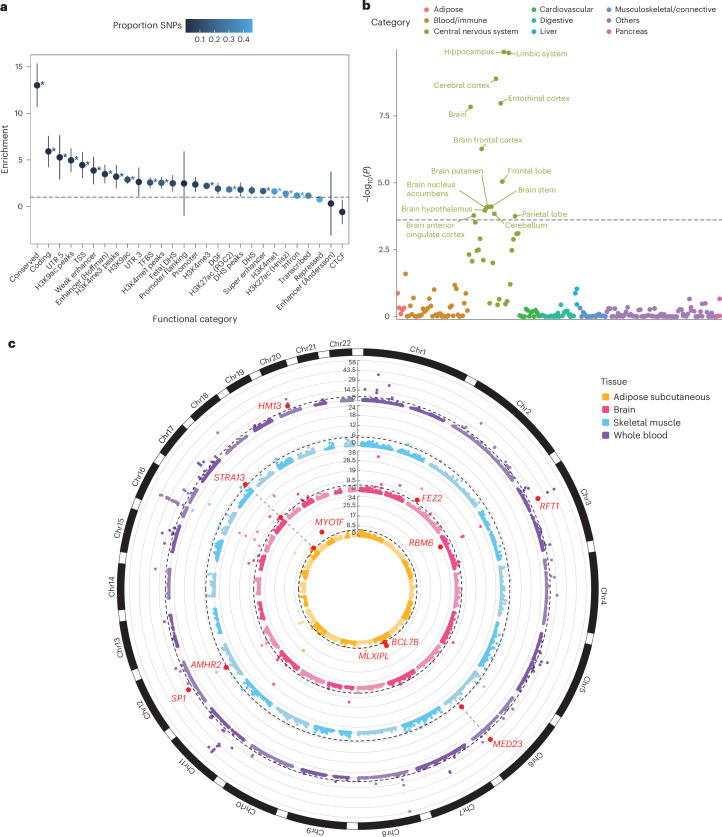
Fig. 3. Post-GWAS analyses of MetS GWAS.
a, Heritability enrichment for functional SNP categories. The x axis represents functional categories, and the y axis represents enrichment. Error bars represent the 95% CIs, calculated as enrichment ± 1.96 × s.e. Asterisks represent significant enrichment after Bonferroni correction (uncorrected P from two-sided enrichment test <9.43 × 10−4), and exact P is available in Supplementary Table 21. b, Tissue-specific enrichment analysis using LDSC-SEG. The x axis represents tissue categories, and the y axis represents the uncorrected −log10(P) from one-sided z test for enrichment in specific tissues. The black dashed line represents the Bonferroni significance threshold of P = 1.02 × 10-4. c, Circos plot62 for gene prioritization using SMR in four tissues. The y axis represents the uncorrected −log10(P) from two-sided χ²-test with 1 degree of freedom for the gene association. Red dots with annotations show replicated genes. The dashed black line represents the Bonferroni significance threshold for the corresponding tissue. Chr, chromosome; CTCF, CCCTC-binding factor; DGF, digital genomic footprint; DHS, DNase I hypersensitivity site; TFBS, transcription factor binding site; TSS, transcription start site; UTR, untranslated region.