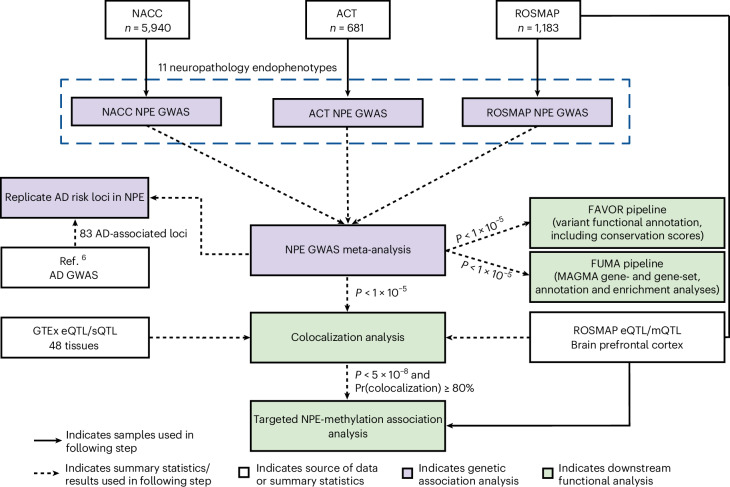
Fig. 1. Overview of GWAS meta-analysis study design.
We performed GWAS meta-analyses of 11 NPEs across three data sources. White boxes represent data sources or summary statistics used in this study. Purple boxes represent individual steps throughout the genetic association analysis, and green boxes represent downstream functional analyses. The first stage of this analysis involved independent GWAS performed in parallel across the NACC neuropathology dataset, the ACT study and the combined ROSMAP. We then performed a meta-analysis using results from each individual GWAS using METAL. Variants reaching a suggestive threshold of association (P ≤ 1 × 10−5) in the meta-analysis were then carried forward for downstream analyses, including functional and colocalization analyses. Variants reaching the genome-wide significant threshold (P ≤ 5 × 10−8) and exhibiting ≥80% colocalization between two NPEs were followed up using existing methylation data to assess the association. All variants reaching genome-wide significance were considered associated with the respective NPE. We also report variants that reached a suggestive threshold (P ≤ 5 × 10−7) or reached the lower suggestive threshold (P ≤ 1 × 10−5) and were in a previously known disease-associated locus. GTEx, Genotype-Tissue Expression Project; QTL, quantitative trait locus; eQTL, expression QTL; sQTL, splicing QTL; mQTL, methylation QTL; AD, Alzheimer’s disease.