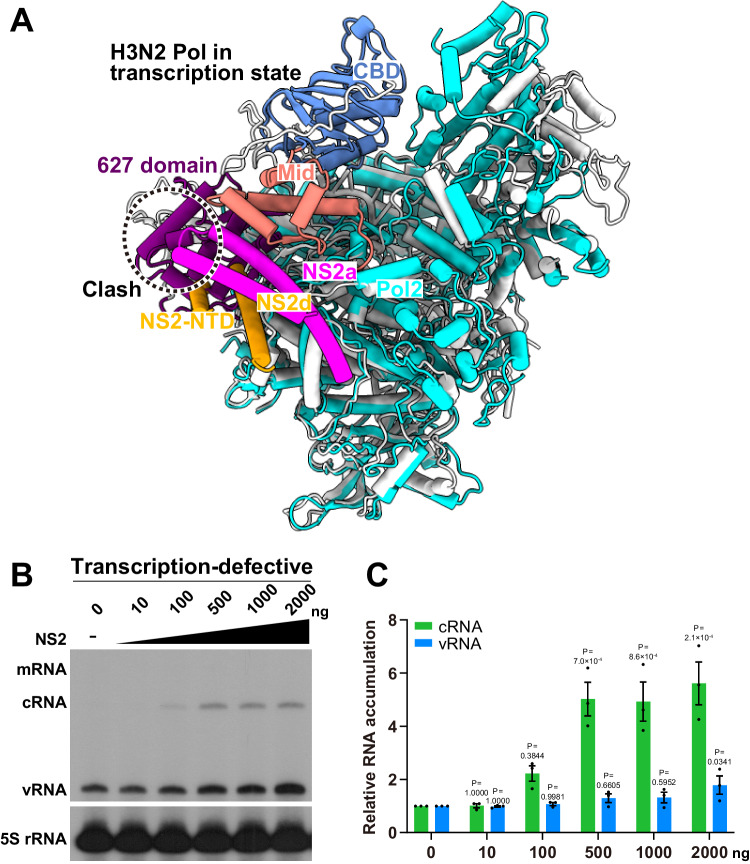
Figure 5. NS2 prevents FluAPol from adopting the transcriptase conformation.
(A) Structural comparison of H3N2 polymerase in the transcription state (PDB ID:6RR7) and a FluAPol protomer from the FluAPol-NS2 hexamer complex. During transcription, the cap binding domain of PB2 and the endonuclease domain of PA should be aligned to enable the cap-snatching process. In the structure of the FluAPol-NS2 complex, binding of NS2 would cause a steric clash with the 627 domain of PB2 and thus prevents FluAPol from adopting the transcriptase conformation. (B) Dose-dependent effect of NS2 on the accumulation of viral RNAs in the transcription-defective (PA-D108A) RNP reconstitution system in HEK 293T cells. (C) Quantification of the effects of NS2 on viral RNA synthesis shown in (B). The graph shows the relative mean intensities of viral cRNA and vRNA normalized to 5S rRNA. The graph shows the mean ± s.e.m. from three biological replicates. Source data are available online for this figure.