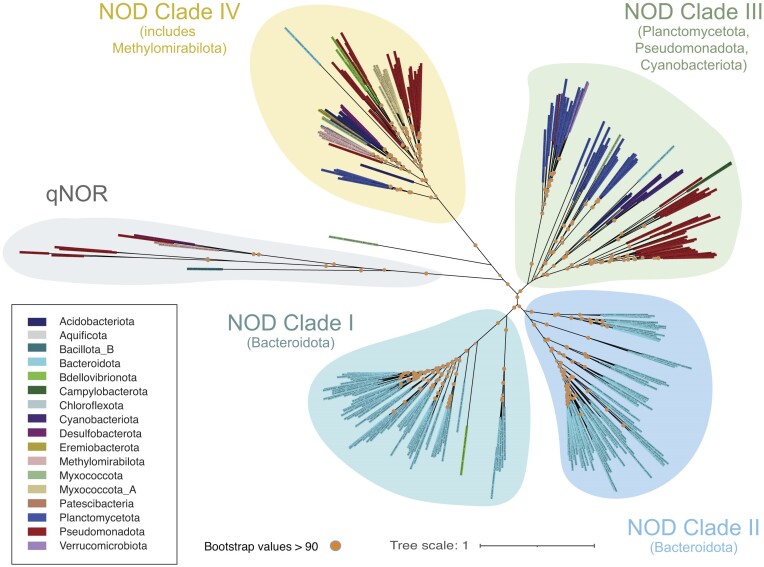
Figure 3.
Phylogenetic diversity and taxonomic distribution of NOD. NOD sequences were retrieved from release 214 of the GTDB (Parks et al. 2022) using an HMM, built with curated NOD sequences (Murali et al. 2022), available on GitHub (https://github.com/ranjani-m/HCO). These sequences were aligned using MUSCLE (Edgar 2004) ), and a phylogenetic tree was inferred using IQ-TREE (Minh et al. 2020) with qNOR sequences as outgroup. A substitution model was identified with IQ-TREE’s ModelFinder, and the tree was validated with 1000 ultrafast bootstraps. The tree was visualized using iTOL, and the label background of each leaf on the tree was colored according to the phylum of the bacteria or archaea that contained the NOD (Supplementary Table 1). The phylum-level classification was made according to GTDB taxonomy. The tree appeared to diverge into four clades, each of which is color-coded here. The protein accession ID of each sequence in the tree according to GTDB is available in Supplementary Table 1.