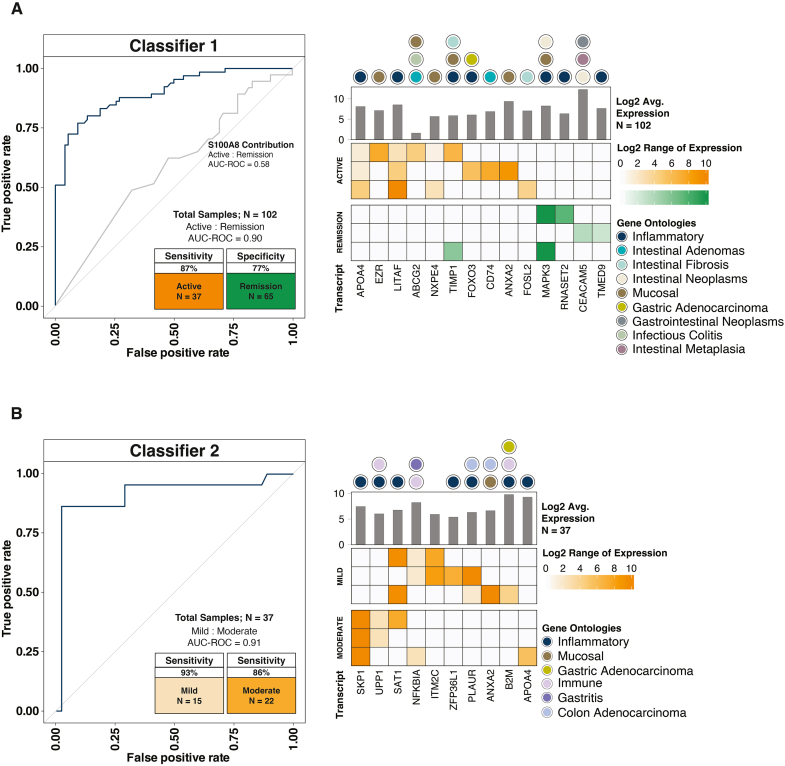
Figure 2.
Machine learning model classifiers show seRNA's ability to approximate disease activity. For each classifier, the top 3 biomarker signatures are shown. Each row in the heatmap indicates a composite biomarker, which is composed of highlighted transcripts in that row. The shade of the highlight indicates transcript expression. Gene ontologies for each transcript are also provided. (A) Classifier 1 predicted subjects with active disease (n = 37) relative to subjects in remission (n = 65) when compared to the Crohn’s Disease Activity Index (CDAI). The receiver operating characteristic (ROC) curve for the RNA calprotectin subunit (S100A8) is shown to provide a surrogate marker for fecal protein calprotectin (ROC area under the curve (AUC) = 0.58). (B) For subjects with active disease, Classifier 2 further parsed those with mild disease (n = 15) relative to those with moderate disease (n = 22) when compared to the CDAI.