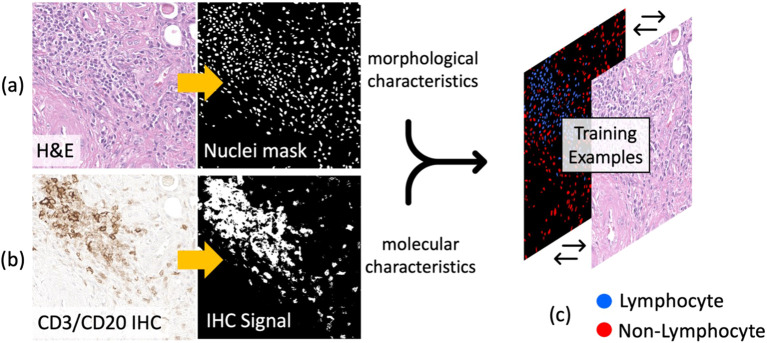
Figure 4.
Automatic lymphocyte labeling to generate deep learning training data at single-cell resolution. (A) Nuclei are first identified and segmented using a pretrained StarDist model. (B) The CD3/CD20 IHC signal (brown stain) is then thresholded and mapped onto the segmented nuclei to identify lymphocytes on H&E. (C) Deep learning training examples are curated by labeling nuclei as either lymphocytes (positive class: overlap between nuclear contour and IHC signal) or non-lymphocytes (negative class: non-overlap between nuclear contour and IHC signal).