To the Editor
Gout is a well-known inflammatory arthritis characterized by sudden, severe pain attacks. It is caused by persistent hyperuricemia and the accumulation of monosodium urate crystals (MSU) in the joint, leading to inflammation and intense pain [1]. To date, we have identified multiple loci associated with serum urate levels [2] and clinically defined gout [1] from genome-wide association studies (GWASs). Recently, after 28 loci were selected from 36 loci identified by the GWAS of serum urate levels in 121,745 Japanese individuals [2], nine loci were reported to be significantly associated with clinically defined gout [3], using the results of the previous gout GWAS [1].
In the present study, to identify further gout loci, we analyzed more selected loci from the results of the trans-ethnic meta-analysis of serum urate levels in 232,092 individuals [2], which discovered 59 statistically significant loci. Of these, 42 urate-related loci were selected for the present association study of clinically defined gout, since the two previous reports [1, 3] identified a total of 17 loci associated with clinically defined gout (ABCG2, CUX2 (ALDH2), SLC2A9, SHLD2/FAM35A, GCKR, NRXN2, SLC17A1, BCAS3, UNCX-MICALL2, BICC1, EMX2-RAB11FIP2, NFAT5, PDZK1, LRP2, PRDM8-FGF5, MLXIPL (BAZ1B), IGF1R). Supplementary Table S1 shows the results of the association analysis of clinically defined gout for 42 candidate loci. These results were obtained by the summary data of our previous gout GWAS [1], in which 3053 male cases with clinically defined gout and 4554 normouricemic male controls (with no gout history and serum urate ≤ 7.0 mg/dl) were analyzed. The level of significance α was set to a p value of < 1.19 × 10–3 (= 0.05/42 with Bonferroni correction).
As shown in Table 1, the present study revealed five loci that were significantly associated with clinically defined gout. Of these five loci, two loci (ORC4 and MYO9A) were identified as novel gout loci, and NRG4 was firstly reported to be associated with clinically defined gout in the present study (Table 1). Two remaining loci, A1CF and MLXIP, were reported in our other previous papers to be related to clinically defined gout [4, 5]. The results of three loci (ORC4, MYO9A, NRG4) were as follows: rs2307394 [a missense variant, p.Asn78Ser, in origin recognition complex subunit 4 (ORC4)] (p value = 2.80 × 10−5; odds ratio (OR): 1.15; 95% confidence interval (CI) 1.08—1.24), rs2957742 [an intronic variant in myosin IXA (MYO9A)] (p value = 3.54 × 10−4; OR: 1.13; 95% CI 1.06—1.21), and rs4886755 [an intronic variant in neuregulin 4 (NRG4)] (p value = 6.83 × 10−4; OR: 1.13; 95% CI 1.05—1.21). More detailed information on the above three loci (ORC4, MYO9A, NRG4) are provided in the Supplementary Discussion. Here, we briefly describe the association of gout and each locus.
Table 1.
Five loci associated with clinically-defined gout identified in the present study
| SNPa | Locus | Chr | Positionb | Gene | Alleles | Trans-ethnic meta-analysis (SU)c | Association analysis (Gout)d | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Riske | Non-risk | Log10BFf | Posterior probability | OR (95%CI) | p valueg | |||||
| rs2307394 | 2q23.1 | 2 | 148,716,428 | ORC4 (ACVR2A) | C | T | 6.72 | 0.480 | 1.15 (1.08, 1.24) | 2.80 × 10–5 |
| rs10994856 | 10q11.23 | 10 | 52,645,248 | A1CF | A | G | 13.23 | 0.024 | 1.31 (1.13, 1.52) | 2.78 × 10–4 |
| rs7953704 | 12q24.31 | 12 | 122,625,992 | MLXIP | G | A | 9.66 | 0.006 | 1.14 (1.06, 1.21) | 2.08 × 10–4 |
| rs2957742 | 15q23 | 15 | 72,302,894 | MYO9A (PKM) | C | G | 8.54 | 0.027 | 1.13 (1.06, 1.21) | 3.54 × 10–4 |
| rs4886755 | 15q24.2 | 15 | 76,298,132 | NRG4 | G | A | 12.31 | 0.010 | 1.13 (1.05, 1.21) | 6.83 × 10–4 |
a dbSNP rs number
b SNP positions are based on NCBI human genome reference sequence Build hg19
c Results of trans-ethnic meta-analysis of SU were obtained from Ref.2 (Nakatochi, et al., Commun Biol, 2019)
d Results of genome-wide meta-analysis of Japanese clinically-defined gout were obtained from Ref.1 (Nakayama, et al., Ann Rheum Dis, 2020)
e Risk allele is defined as a base which increases SU level and gout risk
f Log10 (Bayes’ factor) of > 6 was adopted for a genome-wide significance level (Ref.2)
g The significance level α was set to a p value of < 1.19 × 10–3 (= 0.05/42 with Bonferroni correction)
Loci identified for the first time in clinically-defined gout cases are shown in bold. Of three loci, two loci including ORC4 and MYO9A were identified as novel gout loci
SNP Single nucleotide polymorphism, Chr Chromosome, SU Serum urate, BF Bayes’ factor, OR Odds ratio, 95%CI 95% Confidence interval
We identified rs2307394 of ORC4 as a novel gout locus (2q23.1). ORC4 is one of the origin recognition complexes that binds specific DNA replication origins to initiate DNA synthesis [6]. However, its physiological and pathophysiological functions on urate handling and/or susceptibility to gout remain unclear. Another gene at the same locus (2q23.1), ACVR2A, encodes a receptor protein involved in the signaling of activins, which form part of the transforming growth factor-beta (TGF-β) family. TGF-β signaling mediates a urate-induced pro-inflammatory phenotype in human monocytes [7], suggesting that ACVR2A might play a role in urate handling and gout development. The present study suggests, for the first time, an association between gout and ACVR2A.
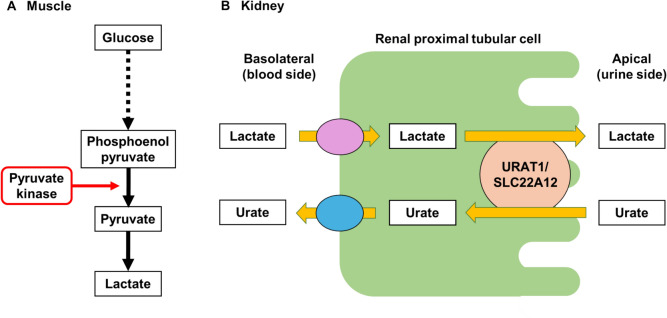
We also identified rs2957742 of MYO9A as a novel gout locus (15q23). MYO9A encodes an atypical myosin that functions as an actin-based molecular motor. It was recently identified as a shared gene signature between rheumatoid arthritis (RA) and colorectal cancer [8]. Given that gout and RA share similar mechanisms such as IL-1β and inflammasome activation, MYO9A might also be associated with gout through a similar inflammatory pathway to that of RA. Another gene at the same locus (15q23), pyruvate kinase M1/M2 (PKM, also known as “pyruvate kinase, muscle”), encodes pyruvate kinase M1/M2, a key enzyme in glycolysis [9]. This enzyme catalyzes the transfer of a phosphoryl group from phosphoenolpyruvate to ADP, producing ATP and pyruvate (Fig. 1A), which results in an increase in lactate. Lactate enhances renal urate reabsorption activity via urate transporter 1 (URAT1), which should lead to an increase in serum urate levels and gout susceptibility (Fig. 1B). This is the first report to suggest an association between gout and PKM.
Fig. 1.
Pyruvate kinase encoded by PKM relates to renal urate reabsorption In the glycolytic system in the tissues such as muscle (A), pyruvate kinase ultimately metabolizes phosphoenolpyruvate to pyruvate, which results in an increase in lactate. In the kidney (B), lactate is known to enhance renal urate reabsorption via urate transporter 1 (URAT1/SLC22A12), which leads to an increase in serum urate levels and gout susceptibility
We for the first time revealed rs4886755 of NRG4 to be a locus (15q24.2) that is significantly associated with clinically defined gout. NRG4 is an adipokine that is primarily secreted by brown adipose tissue. It plays a significant role in regulating energy homeostasis and glucolipid metabolism, and protects against the development of non-alcoholic fatty liver disease (NAFLD) [10]. Oxidative stress, which causes inflammation and hepatic lipotoxicity, is a leading cause of NAFLD. In a previous report, we suggested PNPLA3 to be associated with serum urate levels, since PNPLA3, which is also associated with NAFLD, is related to inflammation and oxidative stress [2]. NRG4 is, therefore, also likely to be associated with serum urate levels and to have protective effects against the inflammation and oxidative stress that causes NAFLD; it thus may have a significant association with gout. The present study is the first to posit an association between gout and NRG4.
In conclusion, the present study indicates the significant association of ORC4, MYO9A, and NRG4 with clinically-defined gout, with ORC4 and MYO9A being identified as novel gout loci. We also suggest that other genes at these three loci—ACVR2A and PKM—could be involved in the pathogenesis of gout. While further studies will be needed on these loci, our approach and findings should lead to a better understanding of the molecular pathogenesis that underlies the development of gout.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We express our sincere thanks to all the participants in this study. Our heartfelt gratitude goes to the members of the Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) for their support. We also thank M. Seki, R. Kikuchi, M. Onishi, Y. Nohara and H. Hanamura (National Defense Medical College) for technical assistance.
Members of the Japan Gout Genomics Consortium (Japan Gout) are: Ken Yamamoto (Department of Medical Biochemistry, Kurume University School of Medicine, Kurume, Japan), Toru Shimizu (Midorigaoka Hospital, Takatsuki, Japan), Hiroshi Ooyama, Keiko Ooyama (Ryougoku East Gate Clinic, Tokyo, Japan), Mitsuo Nagase (Nagase Clinic, Tokyo, Japan), Yuji Hidaka (Akasaka Central Clinic, Tokyo, Japan), Tappei Takada (Department of Pharmacy, The University of Tokyo Hospital, Tokyo, Japan), Kimiyoshi Ichida (Department of Pathophysiology, Tokyo University of Pharmacy and Life Science, Hachioji, Japan), Kenji Wakai, Takashi Tamura (Department of Preventive Medicine, Nagoya University Graduate School of Medicine, Nagoya, Japan), Miki Ueno, Kimiko Hayano (Division of Nursing, National Defense Medical College, Tokorozawa, Japan), Yuzo Takada (Faculty of Medical Science, Teikyo University of Science, Tokyo, Japan), Hiroshi Nakashima, Mitsunobu Tanaka, Noriyuki Yoshioka, Satoko Iwasawa, Masashi Tsunoda (Department of Preventive Medicine and Public Health, National Defense Medical College, Tokorozawa, Japan), Kyoko Morichika, Miho Miyazawa, Mayuko Nakajima, Kazuki Maehara, Mana Kirihara, Yuka Aoyagi, Shin Fujiwara, Yurino Mori, Risa Tanaka, Mio Horie, and Masumi Someya (Department of Integrative Physiology and Bio-Nano Medicine, National Defense Medical College, Tokorozawa, Japan).
Funding
The research conducted by the National Defense Medical College was supported by JSPS KAKENHI [20H00566, 21KK0173, 21H03350, 24K02695, 24K13426, 24K22217, 20K23152, 17H04128, 20H00568, 22H03350, 221S0002, and 16H06279 (PAGS)], the Ministry of Defense of Japan, Grants-in-Aid for Young Scientists of the Japanese Society of Gout and Uric & Nucleic Acids, the Kawano Masanori Memorial Foundation for Promotion of Pediatrics, Suzuken Memorial Foundation, and the Gout and Uric Acid Foundation of Japan. The research conducted by the Nagoya University Graduate School of Medicine was also supported by JSPS KAKENHI [16H06277 (CoBiA) and 22H04923] and Grants-in-Aid for Scientific Research on Innovative Areas (221S0001) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Data availability
Data are available upon reasonable request to the corresponding author.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Data and sample collection for the cohorts participating in the present study were approved by the relevant research ethics committees at the National Defense Medical College (No. 4801) and Aichi Cancer Center (No. H2210001A). All the studies were performed according to the guidelines of the Declaration of Helsinki.
Informed consent
All participants had provided their written informed consent.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yusuke Kawamura, Akiyoshi Nakayama contributed equally to this study.
Contributor Information
Hirotaka Matsuo, Email: matsuo29@gmail.com.
for Japan Gout Genomics Consortium (Japan Gout):
Ken Yamamoto, Toru Shimizu, Hiroshi Ooyama, Keiko Ooyama, Mitsuo Nagase, Yuji Hidaka, Tappei Takada, Kimiyoshi Ichida, Kenji Wakai, Takashi Tamura, Miki Ueno, Kimiko Hayano, Yuzo Takada, Hiroshi Nakashima, Mitsunobu Tanaka, Noriyuki Yoshioka, Satoko Iwasawa, Masashi Tsunoda, Kyoko Morichika, Miho Miyazawa, Mayuko Nakajima, Kazuki Maehara, Mana Kirihara, Yuka Aoyagi, Shin Fujiwara, Yurino Mori, Risa Tanaka, Mio Horie, and Masumi Someya
References
- 1.Nakayama A, Nakatochi M, Kawamura Y, Yamamoto K, Nakaoka H, Shimizu S, et al. Subtype-specific gout susceptibility loci and enrichment of selection pressure on ABCG2 and ALDH2 identified by subtype genome-wide meta-analyses of clinically defined gout patients. Ann Rheum Dis. 2020;79(5):657–65. 10.1136/annrheumdis-2019-216644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakatochi M, Kanai M, Nakayama A, Hishida A, Kawamura Y, Ichihara S, et al. Genome-wide meta-analysis identifies multiple novel loci associated with serum uric acid levels in Japanese individuals. Commun Biol. 2019;2:115. 10.1038/s42003-019-0339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakayama A, Kawamura Y, Nakatochi M, Toyoda Y, Nakajima M, Maehara K et al. Strong genetic effect on gout revealed by genetic risk score from meta-analysis of two genome-wide association studies. Hum Cell. 2024. 10.1007/s13577-024-01138-y.38811494 [Google Scholar]
- 4.Kawaguchi M, Nakayama A, Aoyagi Y, Nakamura T, Shimizu S, Kawamura Y, et al. Both variants of A1CF and BAZ1B genes are associated with gout susceptibility: a replication study and meta-analysis in a Japanese population. Hum Cell. 2021;34(2):293–9. 10.1007/s13577-021-00485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang SJ, Toyoda Y, Kawamura Y, Nakamura T, Nakatochi M, Nakayama A, et al. A meta-analysis of genome-wide association studies using Japanese and Taiwanese has revealed novel loci associated with gout susceptibility. Hum Cell. 2022;35(2):767–70. 10.1007/s13577-021-00665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortega MA, Nguyen H, Ward WS. ORC proteins in the mammalian zygote. Cell Tissue Res. 2016;363(1):195–200. 10.1007/s00441-015-2296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson RD, O’Keeffe GW, Kenny LC. Activin signalling and pre-eclampsia: from genetic risk to pre-symptomatic biomarker. Cytokine. 2015;71(2):360–5. 10.1016/j.cyto.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Zhan ZQ, Huang ZM, Lan QW, Luo YH, Li JX, Zheng YF, et al. Integrated multi-omics analyses revealed the association between rheumatoid arthritis and colorectal cancer: MYO9A as a shared gene signature and an immune-related therapeutic target. BMC Cancer. 2024;24(1):714. 10.1186/s12885-024-12466-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinweha P, Rattanapornsompong K, Charoensawan V, Jitrapakdee S. MicroRNAs and oncogenic transcriptional regulatory networks controlling metabolic reprogramming in cancers. Comput Struct Biotechnol J. 2016;14:223–33. 10.1016/j.csbj.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vulf M, Bograya M, Komar A, Khaziakhmatova O, Malashchenko V, Yurova K, et al. NGR4 and ERBB4 as Promising diagnostic and therapeutic targets for metabolic disorders. Front Biosci (Elite Ed). 2023;15(2):14. 10.31083/j.fbe1502014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request to the corresponding author.