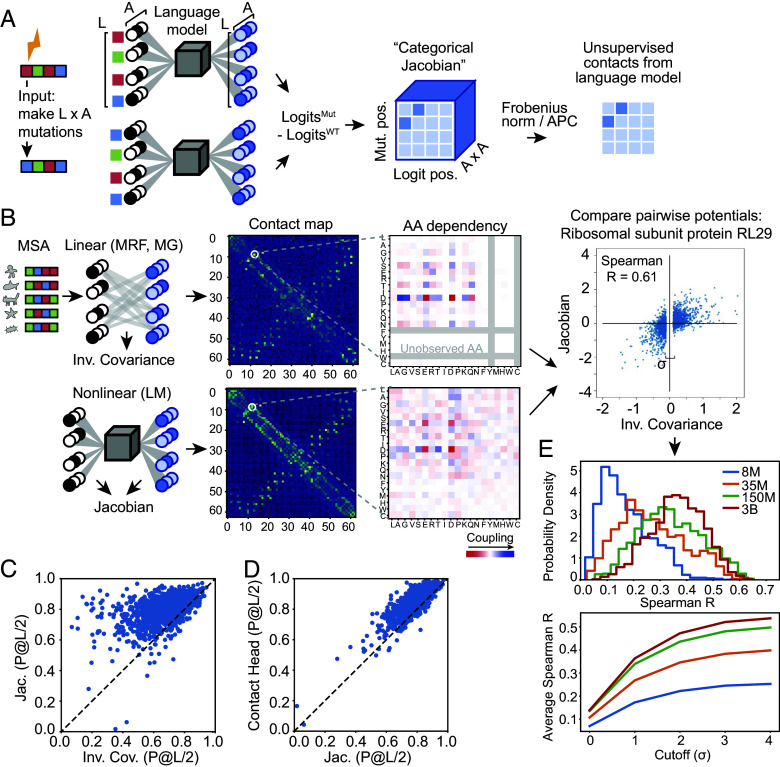
Fig. 4.
Categorical Jacobian is an unsupervised method to extract coevolutionary signal and uniformly evaluate any pLM. (A) Scheme of the categorical Jacobian calculation. Each residue in a sequence of length L is changed to A different types of amino acids, where A is the size of the alphabet (for proteins, ). By computing how the output changes with respect to the input, a matrix of size [L, A, L, A] is obtained. (B) This categorial Jacobian allows for comparing a nonlinear method like ESM-2 and a simple linear method, exemplified here for large ribosomal subunit protein RL29 (UniProt: P0A7M7). We can compare the coevolutionary weights obtained from a linear model, calculated using inverse covariance, and the categorical Jacobian calculated from ESM-2. (C) Contacts calculated from the categorical Jacobian from ESM-2 outperform the inverse covariance calculation from ref. 25 (Average long-range P@L/2 of 0.67 vs. 0.80, respectively). (D) Comparing contact accuracy from the categorical Jacobian and the supervised contact prediction head (Average long-range P@L/2 of 0.80 vs. 0.87, respectively). (E) Correlation between covariation parameters from linear model and ESM-2 Jacobians increase with model size. Top: Distribution of Spearman correlation coefficients between contacts from linear model and ESM-2 Jacobians. Bottom: average Spearman R, varying cutoff for linear model values close to zero. For (C–E), N 1,431 proteins.