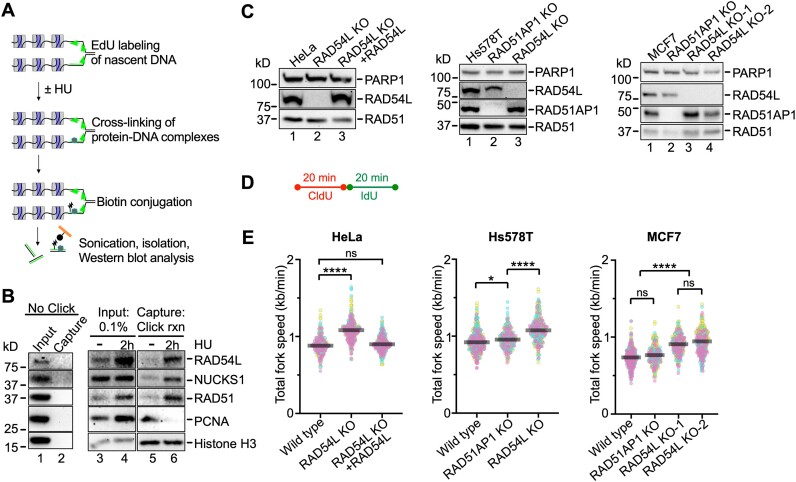
Figure 1.
RAD54L is enriched at stalled replication forks and restrains replication fork progression. (A) Schematic illustration of the iPOND assay. (B) Western blots of the input and iPOND samples probed for proteins, as indicated. NUCKS1 is a RAD54L interacting protein (27). Click rxn, Click reaction; the ‘no Click’ condition represents cells pulsed with EdU and processed without biotin-azide in the Click reaction step. (C) Western blots of HeLa, Hs578T and MCF7 cells and derivatives, as indicated. Loading control: PARP1. (D) Schematic of the protocol for the DNA fiber assay to determine replication speed. (E) SuperPlots (77) with medians of total fork speeds (i.e. CldU + IdU tracts) in unperturbed parental and RAD54L KO cells generated in HeLa, Hs578T, and MCF7 cells. RAD51AP1 KOs and the RAD54L KO ectopically expressing RAD54L-HA (31) are shown for comparison purposes (n = 3; 89–106 fiber tracts/experiment analyzed). Fork speeds were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗∗∗P< 0.0001).