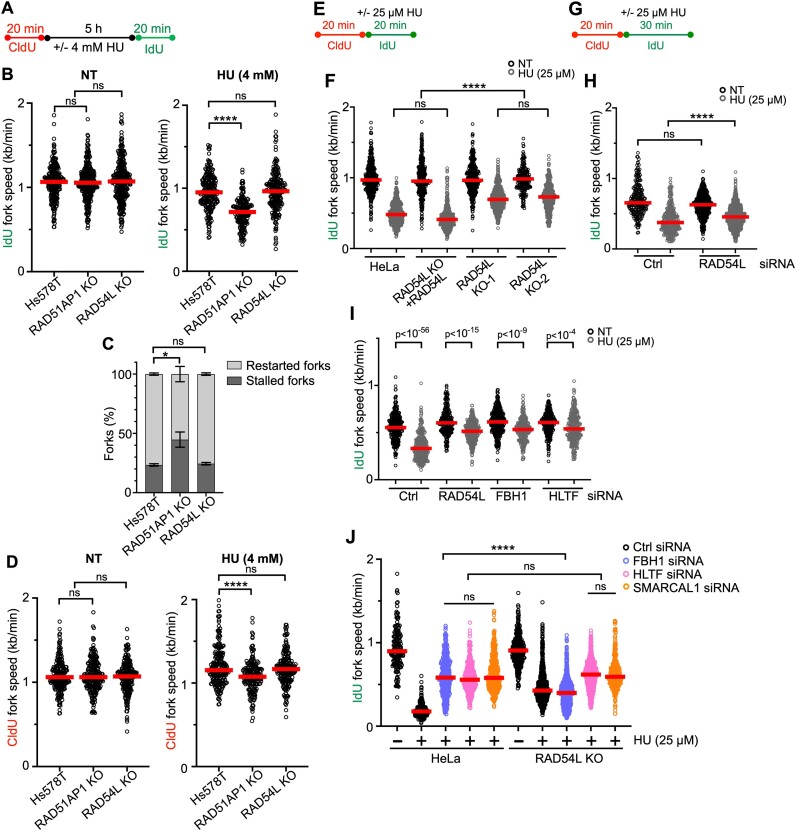
Figure 2.
Loss of RAD54L does not hinder replication restart, leads to faster replication during mild replication stress, and partially restores fork restraint in cells with FBH1 knockdown. (A) Schematic of the DNA fiber assay protocol used to assess replication restart. (B) Dot plot with medians of IdU fork speeds in untreated (NT) or HU-treated parental Hs578T, RAD51AP1 KO, and RAD54L KO cells (n = 3; 65–113 fiber tracts/experiment analyzed). (C) Fractions of stalled and restarted forks after 4 mM HU in the Hs578T cells and derivatives shown in (B). (D) Dot plot with medians of CldU fork speeds in untreated (NT) or HU-treated Hs578T cells and derivatives (n = 3; 65–80 fiber tracts/experiment analyzed). (E) Schematic of the DNA fiber assay protocol used to evaluate the progression of replication forks during mild replication stress in (F). (F) Dot plot with medians of IdU fork speeds in parental HeLa cells, RAD54L KO cells expressing RAD54L KO, and two independently isolated RAD54L KO cell lines treated with or without 25 μM HU during the IdU pulse (n = 3; 82–220 fiber tracts/experiment analyzed). (G) Schematic of the DNA fiber assay protocol used to evaluate the progression of replication forks during mild replication stress in (H), (I) and (J). (H) Dot plot with medians of IdU fork speeds in hTERT RPE-1 cells transfected with Ctrl or RAD54L siRNA and treated with or without 25 μM HU during the IdU pulse (n = 3; 76–293 fiber tracts/experiment analyzed). (I) Dot plot with medians of IdU fork speeds in HeLa cells transfected with control (Ctrl), RAD54L, FBH1 or HLTF siRNA and treated with or without 25 μM HU during the IdU pulse (n = 3; 92–139 fiber tracts/experiment analyzed). (J) Dot plot with medians of IdU fork speeds in HeLa and RAD54L KO cells transfected with Ctrl, FBH1, HLTF or SMARCAL1 siRNA (n = 3; 122–420 fiber tracts/experiment analyzed). All data were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗∗∗P< 0.0001; or as indicated).