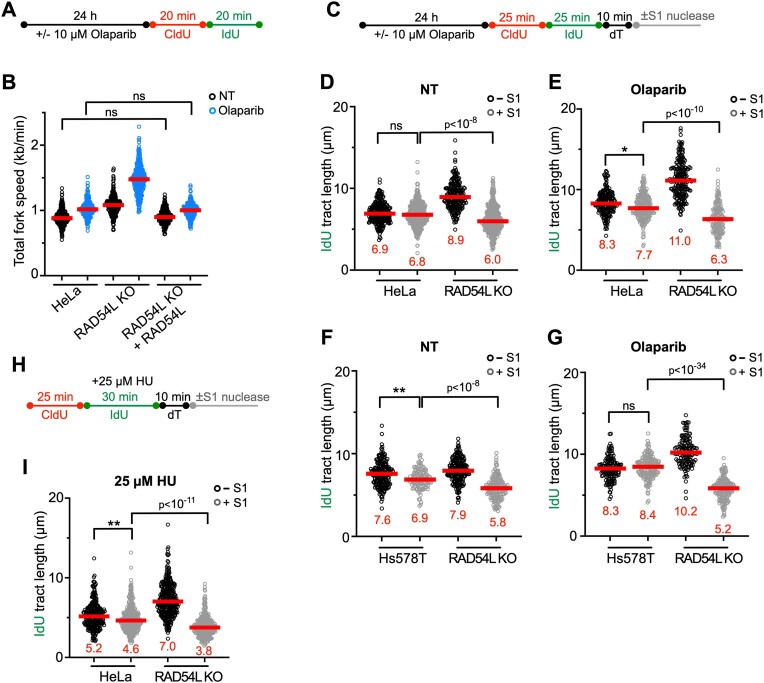
Figure 3.
Loss of RAD54L accelerates replication fork progression and ssDNA gap formation. (A) Schematic of the DNA fiber assay protocol to assess fork progression in (B). (B) Dot plot with medians of total fork speeds (i.e. in CldU + IdU tracts) in HeLa, RAD54L KO and RAD54L KO + RAD54L cells with or without Olaparib (n = 3; 86–100 fiber tracts/experiment analyzed). (C) Schematic of the DNA fiber assay protocol with S1 nuclease in unperturbed and Olaparib-treated cells used in (D)–(G). (D) Dot plot with medians of IdU fiber tract lengths in untreated (NT) HeLa and RAD54L KO cells with or without S1 nuclease (n = 3; 74–149 fiber tracts/experiment analyzed). (E) Dot plot with medians of IdU fiber tract lengths in HeLa and RAD54L KO cells treated with Olaparib and with or without S1 nuclease (n = 3; 60–80 fiber tracts/experiment analyzed). (F) Dot plot with medians of IdU fiber tract lengths in untreated (NT) Hs578T and RAD54L KO cells with or without S1 nuclease (n = 3; 42–70 fiber tracts/experiment analyzed). (G) Dot plot with medians of IdU fiber tract lengths in Hs578T and RAD54L KO cells treated with Olaparib and with or without S1 nuclease (n = 3; 47–63 fiber tracts/experiment analyzed). (H) Schematic of the DNA fiber assay protocol with S1 nuclease in HU-treated cells used in (I). (I) Dot plot with medians of IdU fiber tract lengths in HU-treated HeLa and RAD54L KO cells with or without S1 nuclease (n = 2; 136–169 fiber tracts/experiment analyzed). All data were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗P< 0.01; or as indicated).