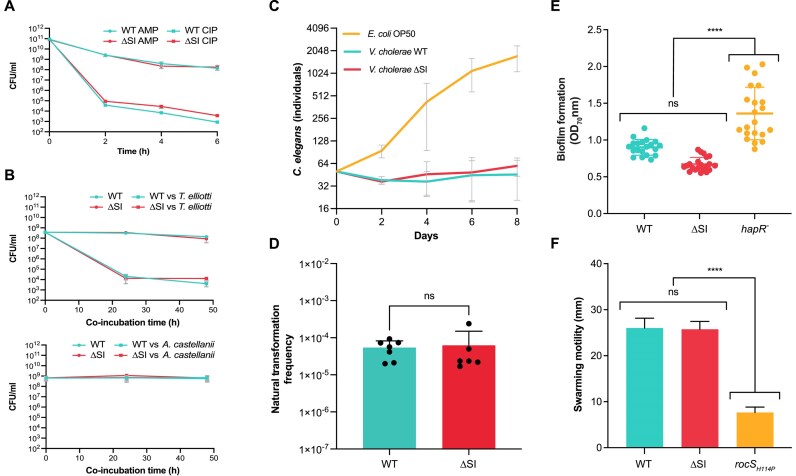
Figure 8.
(A) Antibiotic persistence assay of V. cholerae WT and ΔSI mutant challenged with 10-fold the MIC of ampicillin (AMP) and ciprofloxacin (CIP), individually. CFUs/ml are plotted against time. Error bars indicate standard deviations from three independent biological replicates. (B) V. cholerae WT and ΔSI survival under predation by T. elliotti (MOI 1:10000) or A. castellanii (MOI 1:500). The number of surviving bacteria, incubated either in the absence (circles) or presence (squares) of ciliates/amoeba, was quantified by CFU counting after 0–48 h of co-incubation. Error bars indicate standard deviations from at least three independent biological replicates. (C) Virulence assay of V. cholerae WT and ΔSI mutant in a C. elegans model. E. coli OP50 was used as a negative control. Error bars indicate standard deviations from three independent biological replicates. (D) Natural competence assay of V. cholerae WT and ΔSI mutant. Transformation frequencies are given on the Y-axis and were calculated as the number of CFUs growing in kanamycin over the total CFUs. Bar charts show the mean ± s.d. from biological replicates (individual dots) from at least three independent experiments. (E) Biofilm formation of V. cholerae WT and ΔSI mutant. A V. cholerae hapR (frameshift) mutant was included as a positive control of biofilm formation. Data from at least 20 independent colonies is represented. (F) Swarming motility assay of V. cholerae WT and ΔSI mutant. A V. cholerae rocS (H114P) mutant was included as a non-motile control strain. Error bars indicate standard deviation of at least three biological replicates. P-values (****) were calculated by pairwise comparison using the one-way ANOVA test (P<0.0001). Ns: not significant.