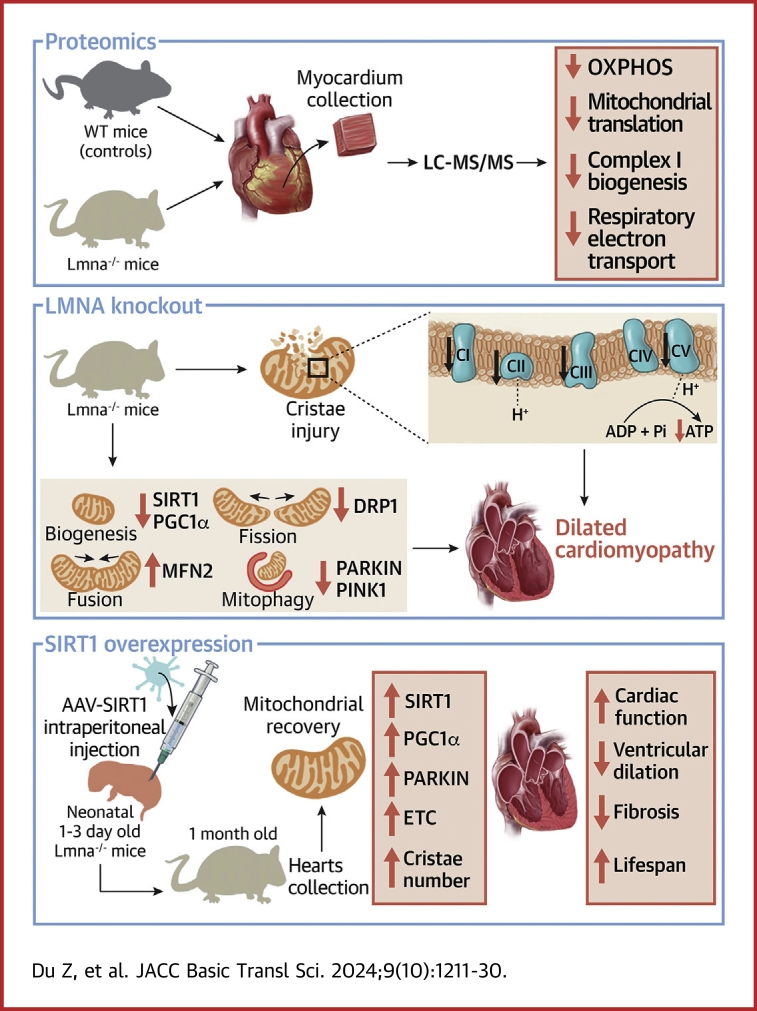
Visual Abstract
Key Words: dilated cardiomyopathy, LMNA, mitochondrial bioenergetics, proteomics, SIRT1
Highlights
-
•
Lamin A/C deficiency leads to aberrant mitochondrial structure and function.
-
•
Mitochondrial biogenesis damage, fission and fusion imbalance, and mitophagy defects are present in Lmna–/– mice hearts, and SIRT1 has been down-regulated before the onset of DCM.
-
•
AAV-mediated SIRT1 overexpression alleviates mitochondrial injury, cardiac systolic dysfunction, ventricular dilation and fibrosis, and prolonged lifespan in Lmna–/– mice.
-
•
LMNA maintains mitochondrial bioenergetics through the SIRT1-PARKIN axis.
Summary
Dilated cardiomyopathy (DCM) is associated with high mortality despite advanced therapies. The LMNA gene encodes lamin A/C and is the second most frequently mutated gene associated with DCM, for which therapeutic options are limited. Here we generated Lmna–/– mice and found they exhibited cardiac dysfunction at the age of 1 month but not at 2 weeks. Proteomics showed down-regulation of mitochondrial function–related pathways in Lmna–/– hearts. Moreover, early injured mitochondria with decreased cristae density and sirtuin 1 (SIRT1) down-regulation were observed in 2-week-old Lmna–/– hearts. Adenoviral overexpression of SIRT1 in lamin A/C knockdown neonatal rat ventricular myocytes improved mitochondrial oxidative respiration capacity. Adeno-associated virus–mediated SIRT1 overexpression alleviated mitochondrial injury, cardiac systolic dysfunction, ventricular dilation, and fibrosis, and prolonged lifespan in Lmna–/– mice. Mechanistically, LMNA maintains mitochondrial bioenergetics through the SIRT1-PARKIN axis. Our results suggest that targeting the SIRT1 signaling pathway is expected to be a novel therapeutic strategy for LMNA mutation–associated DCM.
The LMNA gene encodes the nuclear envelope protein lamins A and C (lamin A/C), 2 major isoforms of A-type lamins arising by alternative splicing.1 In addition, pathogenic LMNA variants have been shown to bring about a spectrum of clinical disorders with heterogeneous manifestations, collectively known as laminopathies.2 It has been reported that 5% to 10% of idiopathic dilated cardiomyopathy (DCM) and 5% of familial DCM are related to LMNA mutations.3,4 Patients with LMNA mutations–associated DCM (LMNA-DCM) have a worse prognosis due to a high risk of premature death and pump failure,4 but specific therapeutic modalities remain limited.
Lamin A/C is pivotal to various biological processes, including gene expression regulation, mechanical sensing and force transmission, DNA replication, cytoplasmic transport, and signal transduction.5 Multiple pathways, such as extracellular signal-regulated kinase/c-Jun N-terminal kinase/mitogen-activated protein kinase p38 alpha, mammalian target of rapamycin, MKL/myocardin-like protein 1/serum response factor, forkhead box protein O, and cellular tumor antigen p53,6, 7, 8, 9, 10, 11, 12, 13 have been found to be involved in the pathogenesis of LMNA-DCM. Furthermore, it has been shown that platelet-derived growth factor signaling mediates the LMNA mutations-associated cardiac arrhythmias.14 Recently, RNA sequencing studies indicated impaired oxidative phosphorylation (OXPHOS) in Lmna–/– hearts.12,15 Moreover, some LMNA mutations have been found to contribute to mitochondrial uncoupling in mouse myoblasts.16 Accumulation of progerin, a toxic truncated prelamin A protein causing premature aging, significantly induced mitochondrial dysfunction.17,18 However, it remains unknown to what extent LMNA impairment leads to mitochondrial bioenergetics damage in cardiomyocytes as well as the underlying mechanism.
The sirtuin protein family are class III nicotinamide-adenine dinucleotide, oxidized form (NAD+)-dependent histone deacetylases, among which sirtuin 1 (SIRT1) was the first to be discovered and plays multiple roles in senescence, apoptosis, calorie restriction, and metabolic homeostasis.19 SIRT1 deacetylates peroxisome proliferator–activated receptor gamma coactivator 1-alpha (PGC1α), thereby promoting mitochondrial biogenesis and energetic metabolism.20,21 In particular, SIRT1 also reportedly regulates mitophagy, a specialized autophagic approach eliminating injured mitochondria, by enhancing the expression of PTEN-induced kinase 1 (PINK1) and its cytosolic ubiquitin ligase PARKIN, which is protective in myocardial infarction and aging.22, 23, 24
The link between lamin A/C and sirtuins has been reported. In a laminopathy-based progeria mice model, lamin A acts as a SIRT1 activator, and SIRT1-targeting therapy mitigated progeroid features and extended the lifespan.25 Another study revealed a vital role for lamin A in endogenously activating sirtuin 6, which promoted beneficial effects on sirtuin 6–mediated DNA repair in Hutchinson-Gilford progeria syndrome.26 In addition, vascular endothelium–targeted Sirt7 overexpression significantly ameliorated endothelial dysfunction and premature death in progeria mice.27 Nonetheless, it remains unknown whether sirtuin protein plays a role in cardiac laminopathy.
In the present study, we generated Lmna–/– mice using the CRISPR/Cas9 technique and performed untargeted proteomic studies. Mitochondrial dysfunction and SIRT1 down-regulation were identified in Lmna–/– hearts before the DCM phenotype was evident. Overexpression of SIRT1 ameliorated LMNA deletion–induced mitochondrial injury and cardiac dysfunction partially by PARKIN up-regulation. These findings suggest that the SIRT1-PARKIN axis is a novel target for therapeutic interventions for treating LMNA-DCM.
Methods
Animal Experiments
All animal experiments were performed according to the guidelines from the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care Committee of Shanghai Jiao Tong University School of Medicine. Mouse LMNA transcript (consisting of 12 exons located in murine chromosome 1) was identified using the ENSEMBL database (transcript identifier: ENSMUST00000029699.12). LMNA heterozygous (Lmna+/–) mice were generated under a C57BL/6J background using the CRISPR/Cas9 technique through non-homologous recombination repair introducing mutation to knock out exons 8 to 11 (Supplemental Figure 1A). Guide RNA (gRNA) targeting exon 8 (gRNA1 sequence: GAAGATGCATGATATAGGGA TGG), gRNA targeting exon 11 (gRNA2 sequence: GGTGGAGCGGGAGCCCAGGT GGG), and Cas9 messenger RNA (mRNA) were obtained from in vitro transcription. Lmna+/– mice were hybridized with wild-type (WT) C57BL/6J mice and further propagated to obtain LMNA knockout (Lmna–/–) mice. Positive Lmna–/– mice were identified by using polymerase chain reaction analysis (Supplemental Figure 1B). Primer 1 sequence was as follows: ‘GGCGGTAGAGGAAGTCGATG’, forward; ‘TCCTGCTGTAGGGCAGAGAT’, reverse (679 bp). Primer 2 sequence was as follows: ‘GGCGGTAGAGGAAGTCGATG’, forward, ‘GAGGCCAACTTCTCCACTCC’, reverse (353 bp). Mice were sacrificed by using an intraperitoneal injection of 1% pentobarbital sodium (100 mg/kg) for further analysis. Both male and female mice were used in this study.
Echocardiogram
Echocardiographic analysis was performed by using a high-resolution ultrasound device (MyLab Touch, Esaote SpA; linear array probe; frequency 18-22 MHz). Mice were anesthetized by isoflurane (2.5%) via inhalation and placed on a heated platform (37 °C) for accurate gathering of echocardiographic data. The left ventricular systolic diameter and diastolic diameter were measured at parasternal long-axis views. The left ventricular end-systolic volume (LVESV), left ventricular end-diastolic volume (LVEDV), left ventricular ejection fraction (LVEF), and fractional shortening (FS) were calculated by using the Teichholz method to assess ventricular function. The operators were blinded to the animal genotypes.
Histological Analysis
After intraperitoneal injection with a lethal dose of pentobarbital sodium (100 mg/kg), mice were transcardially perfused with 20 mL of saline solution. Mice hearts were then harvested and immediately fixed in 4% paraformaldehyde at room temperature, followed by dehydration and paraffin embedding. Paraffin blocks were sliced with a thickness of 4 μm and stored for further use. The sections were stained with hematoxylin and eosin or Masson’s trichrome for assessment of heart morphology and fibrosis according to the manufacturer’s protocol.
Transmission Electron Microscopy
Fresh myocardial tissue specimens of the mice were cut into 1 mm3 pieces and immediately fixed in 2.5% glutaraldehyde fixation solution. The tissues were preserved at 4 °C, followed by fixation with 1% osmic acid away from light, and washed with 0.1 M phosphate buffer. After dehydration, the samples were embedded in ethoxyline resin and sliced into 60 to 80 nm sections, followed by staining with 2% uranyl acetate saturated alcohol solution and 2.6% lead citrate solution. The sections were observed by using an HT7800 transmission electron microscope (Hitachi). The images were quantitatively analyzed according to a standardized approach using ImageJ software version 1.8 (National Institutes of Health).28 In addition, mitochondria were manually divided into 3 categories based on cristae density: healthy (cristae density >90%), mildly injured (cristae density between 50% and 90%), and severely injured (cristae density <50%), as previously reported.29 More than 50 mitochondria were randomly selected from each sample for quantitative analysis.
Proteomic Analysis
Proteomics was performed by using nanoElute (Bruker) coupled to a timsTOF Pro (Bruker). Raw files were analyzed against the Mouse SwissProt FASTA database (UP000000589) with DIA-NN (version 1.8.1) (details are provided in the Supplemental Methods).
Adeno-Associated Virus Construction and Injection
The adeno-associated virus (AAV) serotype 9 for SIRT1 overexpression was designed and constructed by OBiO Technology. Briefly, plasmids pAAV-CMV-3×FLAG-P2A-mNeonGreen-tWPA or pAAV-CMV-Sirt1-3×FLAG-P2A-mNeonGreen-tWPA were transfected into 293T cells. At 72 hours following transfection, cells were collected and lysed by using a freeze-thaw method. The virus particles were purified through iodiol gradient ultracentrifugation. AAV was intraperitoneally injected to neonatal mice aged 1 to 3 days at a dose of 1.5 × 1011 vg.
Real-Time Quantitative Polymerase Chain Reaction Analysis
Total RNA was extracted from homogenized myocardial tissue using TRIzol (#15596026; Invitrogen), chloroform, and isopropyl alcohol according to a standard protocol and reverse-transcribed to complementary DNA with a gDNA digester and 2×Hifair Ⅱ SuperMix plus (#11123ES10; Yeasen). Complementary DNA transcripts were mixed with SYBR Green I with UDG Mix (#TSE203; Tsingke) and quantified by using the QuantStudio 6 Flex System (Thermo Fisher Scientific). For normalization, 18S ribosomal RNA or glyceraldehyde 3-phosphate dehydrogenase was used. Primers used in this study are listed in Supplemental Table 2.
Western Blot Analysis
Total protein was extracted from myocardial tissue or cells by radioimmunoprecipitation assay containing phenylmethanesulfonyl fluoride and phosphatase inhibitor. The supernatant was denatured with loading buffer at 100 °C for 7 minutes. For mitochondrial respiratory chain proteins, the supernatant was set to boiling at 50 °C. A total of 40 μg protein was separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis and transferred onto 0.45 μm polyvinylidene difluoride membranes, followed by block within 5% skim milk. Membranes were incubated with primary antibodies at 4 °C overnight. After washing thrice in tris-buffered saline + Tween 20, membranes were incubated with either horseradish peroxidase–labeled Goat Anti-Rabbit or Anti-Mouse IgG (Beyotime). Western blotting signals were detected with ECL Reagent (#SQ202; Epizyme) using the GeneGnome XRQ System (Syngene). Expression quantification was performed by using ImageJ. The antibodies are listed as follows:
OXPHOS (1:2000, #ab110413; Abcam), lamin A/C (1:500, #sc-376248; Santa Cruz), SIRT1 (1:1000, #9475; Cell Signaling Technology), PGC1α (1:1000, #A20995; ABclonal), dynamin-related protein 1 (DRP1) (1:1000, #8570; Cell Signaling Technology), mitofusin 2 (MFN2) (1:1000, #9482; Cell Signaling Technology), PARKIN (1:500, #14060-1-AP; Proteintech), PINK1 (1:1000, #23274-1-AP; Proteintech), and β-actin (1:2000, #66009-1-Ig; Proteintech).
Adenosine Triphosphate Assay
The adenosine triphosphate (ATP) level was measured by using the ATP assay kit (#S0026; Beyotime). Briefly, ATP lysate was added at a ratio of 200 μL lysate per 20 mg of tissue and homogenized, followed by centrifugation at 12,000g for 5 minutes. Supernatant was mixed with ATP test solution and added to the nontransparent 96-well plate for measurement of the relative light unit value by using SpectraMax Paradigm Multi-Mode Detection (Molecular Devices). The ATP level were corrected by tissue total protein content.
Isolation and Culture of Neonatal Rat Ventricular Myocytes
Neonatal rat ventricular myocytes (NRVMs) were isolated from neonatal Sprague Dawley rats aged 1 to 3 days according to a standard protocol. Briefly, rats were anesthetized by using 2.5% isoflurane and were then decapitated. The hearts were separated from the chest cavity and cleaned with Hanks’ balanced salt solution to remove the residual blood, cut into 1 to 2 mm3 tissue pieces, and digested overnight in 0.025% trypsin at 4 °C. After the trypsin was replaced by 0.08% type Ⅱ collagenase (#C6885; Sigma-Aldrich), the tissues were digested in a 37 °C water bath for 8 minutes and the process repeated 4 to 5 times. The supernatant was collected and neutralized with Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium containing 10% fetal bovine serum (FBS). Cardiomyocytes were separated by centrifugation at 1,500 rpm and resuspended in 10% FBS/DMEM/F12 medium; they were then purified by differential adherence after 1.5 hours’ culture in an incubator with 5% carbon dioxide. The next day, the medium was replaced by 10% FBS/DMEM/F12 medium, and rhythmically beating NRVMs were obtained for further experiments.
Small Interfering RNA Transfection
NRVMs were transfected with small interfering RNA (siRNA) synthesized by GenePharma. Briefly, when NRVMs reached 70% to 90% confluence and beat rhythmically, cells were transfected with siRNA using Lipofectamine RNAiMAX (#13778150; Thermo Fisher Scientific) in DMEM/F12 medium. Six to eight hours later, the 10% FBS/DMEM/F12 medium was added, and cells were placed in an incubator with 5% carbon dioxide at 37 °C for 72 hours until use. The siRNA sequence targeting rat LMNA is ‘GCUGGUGGAGAUUGAUAAUTT’, forward; and ‘AUUAUCAAUCUCCACCAGCTT’, reverse. The siRNA sequence targeting rat PRKN is ‘CUUUGAACCUGAUCACCAATT’, forward; and ‘UUGGUGAUCAGGUUCAAAGTT’, reverse.
Adenovirus Infection
SIRT1-3×FLAG-eGFP adenovirus vector was constructed and purchased from HanBio Technology. NRVMs were infected with SIRT1-3×FLAG-eGFP or enhanced green fluorescent protein (eGFP) adenovirus in serum-free medium at a multiplicity of infection of 20. Six to eight hours later, the medium was switched to 10% FBS/DMEM/F12 medium. Cells were then incubated at 37 °C for 72 hours for further use.
Cell Bioenergetic Profiling
The Cell Mito Stress Test was performed by using the Seahorse XFe96 extracellular flux analyzer (Agilent Technologies) to monitor the mitochondrial oxygen consumption rate (OCR). Briefly, the steps are as follows: cells were seeded in the microplate at a density of 104 each well and cultured overnight at 37 °C. After transfection or premedication, cells were cultured in base medium containing 10 mM glucose, 1 mM pyruvate, and 2 mM glutamine, followed by maintenance in a carbon dioxide–free incubator at 37 °C for 1 hour. The compounds were automatically injected according to the sequence of 1.5 μM oligomycin, 3 μM FCCP, and 0.5 μM rotenone/antimycin A, when the OCR was continuously monitored. After the test, cells were stained with Hoechst Living Cell Stain Solution (#C1027; Beyotime) and counted by using Cytation 5 (Agilent Technologies) for OCR normalization.
Live Cell Imaging
After 72 hours of NRVM transfection, the medium was discarded and displayed by 150 nM tetramethylrhodamine methyl ester (#I34361; Thermo Fisher Scientific) for 30 minutes or 5 μM MitoSOX (#M36008; Thermo Fisher Scientific) for 10 minutes or 200 nM MitoTracker Red (#M22425; Thermo Fisher Scientific) for 30 minutes diluted by Hanks’ balanced salt solution at 37 °C. Cells were observed using Cytation 5 or OMX imaging (GE Healthcare). To avoid biases, the same exposure parameters (ie, illumination intensity, integration time, camera gain) were kept between samples in different groups during signal acquisition. The fluorescence intensity was quantified by using ImageJ.
As for mitochondrial network analysis, images were processed by using the Mitochondrial Network Analysis method as previously described.30,31 Four computed parameters were used in this study: footprint (total area of mitochondrial signal), individuals (punctate, rod, and large mitochondria), networks (mitochondrial networks with at least one junction), and mean branches per network (mean number of branches per mitochondrial network).
Statistical Analysis
All data are expressed as mean ± SD. The Shapiro-Wilk test was used to evaluate the normality of the data. If data were normally distributed, 2 groups were compared by using Student's t-test, and ≥3 groups were compared by using one-way analysis of variance with a Tukey post hoc test for multiple pairwise comparisons. Otherwise, the Mann-Whitney U test (2 groups) or the Kruskal-Wallis test (≥3 groups) was used with the Dunn post hoc test for multiple pairwise comparisons. Kaplan-Meier curves were used to evaluate survival, with groups compared by using the Breslow generalized Wilcoxon test.32,33 Detailed statistical methods for mass spectrometry data are described in the Supplemental Methods.
Statistical analyses were performed by using GraphPad Prism 8.0 software (GraphPad Software) or R 4.2.2 (R Foundation for Statistical Computing) and RStudio version 2023.03.0+386 (Posit PBC). P values <0.05 were considered statistically significant.
Results
One-Month-Old but not 2-Week-Old Lmna–/– Mice Exhibit Cardiac Dysfunction
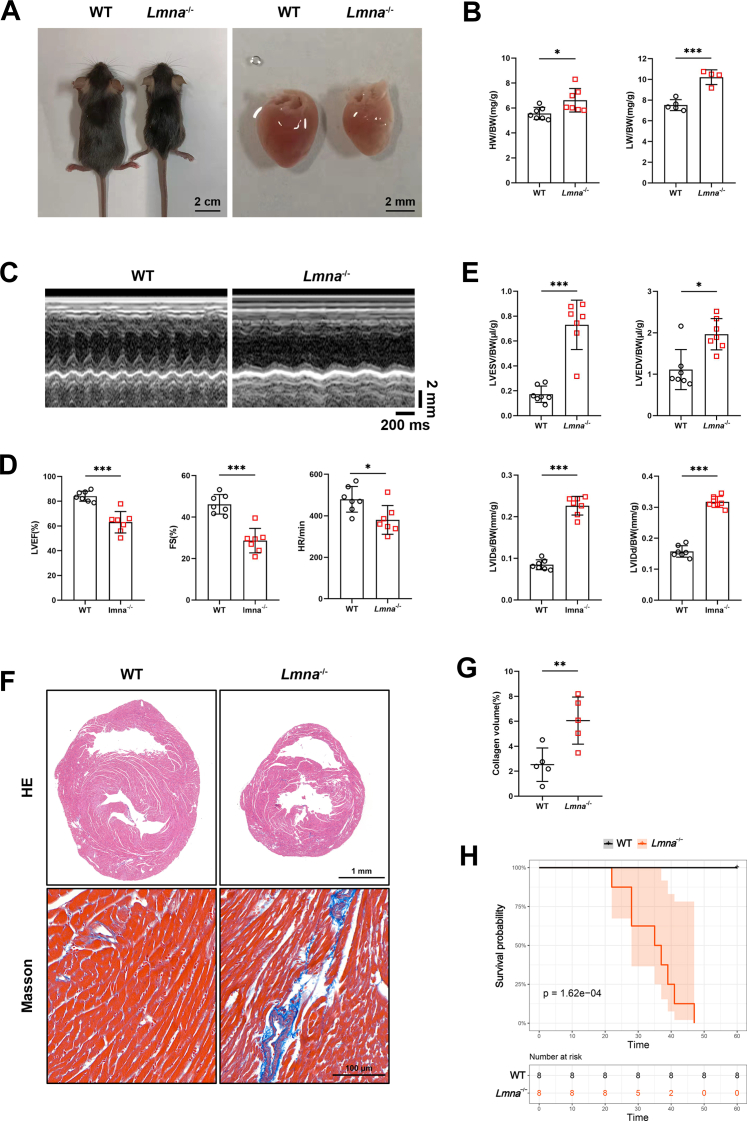
Lmna–/– mice were generated by a deletion from exon 8 to the middle of exon 11 of the Lmna gene using the CRISPR/Cas9 technique (Supplemental Figures 1A and 1B). Consistent with previous studies,34,35 1-month-old Lmna–/– mice had a smaller body and heart size/weight, but with higher heart weight (HW) to body weight (BW) ratio (HW/BW), and lung weight (LW) to BW ratio (LW/BW), compared with WT mice (Figures 1A and 1B, Supplemental Figure 1C). Echocardiography revealed significantly reduced LVEF, FS, and heart rate in Lmna–/– mice (Figures 1C and 1D). When normalized to BW, LVESV, LVEDV, left ventricular internal diameter at end-systole (LVIDs), and left ventricular internal diameter at end-diastole (LVIDd) of Lmna–/– hearts were overtly higher than in their littermate WT controls (Figure 1E). Histological analysis also showed that Lmna–/– hearts had an enlarged heart cavity and thinner ventricular wall despite a smaller heart size (Figure 1F). Masson staining also revealed increased cardiac fibrosis in Lmna–/– hearts (Figures 1F and 1G). These results were further supported by elevated mRNA levels of Nppa, Nppb, and Myh7, which are markers of heart failure, and Col1a1, but not Col3a1, which are markers of fibrosis, in Lmna–/– hearts (Supplemental Figure 1D).
Figure 1.
One-Month-Old Lmna–/– Mice Exhibit Cardiac Dysfunction
(A) Representative exteriors (scale bar: 2 cm) and cardiac appearances (scale bar: 2 mm) of 1-month-old wild-type (WT) and Lmna–/– mice. (B) Heart weight (HW)/body weight (BW) (n = 7) and lung weight (LW)/BW (WT, n = 5; Lmna–/–, n = 4) of WT and Lmna–/– mice. (C) Representative echocardiography of WT and Lmna–/– mice. (D) Left ventricular ejection fraction (LVEF), fractional shortening (FS), and heart rate (HR) in WT and Lmna–/– mice (n = 7). (E) Left ventricular end-systolic volume (LVESV)/BW, left ventricular end-diastolic volume (LVEDV)/BW, left ventricular end-systolic diameter (LVIDs)/BW, and left ventricular end-diastolic diameter (LVIDd)/BW in WT and Lmna–/– mice (n = 7). (F) Representative hematoxylin and eosin (HE) staining (scale bar: 1 mm) and Masson’s trichrome staining (scale bar: 100 μm) images of the ventricular transverse section of WT and Lmna–/– mice. (G) Quantification of the percentage of collagen in myocardial tissue of WT and Lmna–/– mice (n = 5). (H) Kaplan-Meier curves of WT and Lmna–/– mice showing a median survival time of 36 days for Lmna–/– mice (n = 8, Breslow generalized Wilcoxon test). Data are presented as mean ± SD. Data in B, D, E (LVIDs/BW, LVIDd/BW), and G were compared by using Student’s t-test, and data in E (LVESV/BW, LVEDV/BW) were compared by using the Mann-Whitney U test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
The median postnatal lifespan of Lmna–/– mice was 36 days (Figure 1H). In contrast, 2-week-old Lmna–/– mice had similar cardiac function and structure to WT controls as shown by echocardiography and histological analysis (Supplemental Figure 2). These results indicate that 1-month-old Lmna–/– mice manifest severe DCM phenotype.
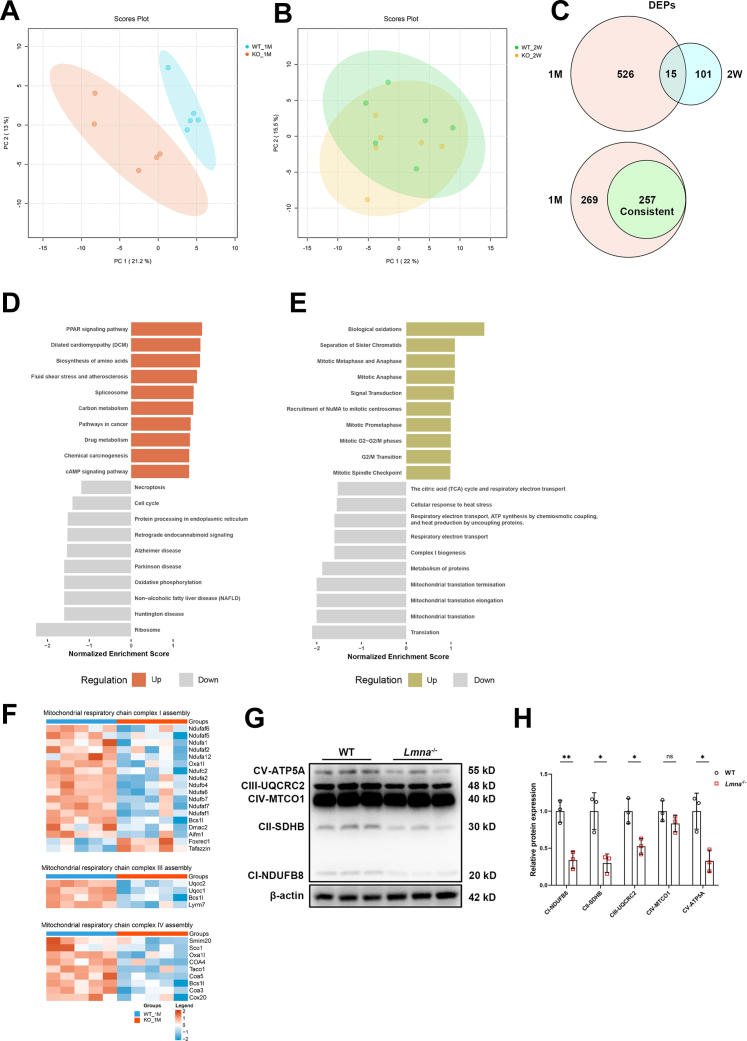
Proteomics Reveals Down-regulated Proteins Related to Mitochondrial Function in Lmna–/– Mice Hearts
Because the cardiac dysfunction in Lmna–/– mice was observed at 1 month but not at 2 weeks, proteomics analysis of Lmna–/– and WT heart tissues was performed at these 2 time points to explore the underlying mechanisms of LMNA deletion–induced DCM. A total of 6,963 proteins were identified, with 6,240 proteins quantified in ≥60% of samples (Supplemental Table 1). Principal component analysis revealed that the protein clustering of Lmna–/– and WT myocardial tissue could be clearly distinguished at 1 month of age but not at 2 weeks (Figures 2A and 2B). Volcano plots illustrated all the differentially expressed proteins (DEPs) with fold change ≥1.3 (Supplemental Figures 3A and 3B). Overall, a total of 526 proteins were differently expressed between the Lmna–/– and WT groups at 1 month, whereas only 101 were differently expressed at 2 weeks. In addition, 257 of the 526 DEPs in 1 month were consistent with the trend of changes seen at 2 weeks, most of which had a higher fold change (Figure 2C, Supplemental Figure 3C). Gene Set Enrichment Analysis revealed that enriched Kyoto Encyclopedia of Genes and Genomes pathways, including ribosome, OXPHOS, and protein processing in the endoplasmic reticulum, were significantly down-regulated, whereas peroxisome proliferator–activated receptor signaling pathway and biosynthesis of amino acid were up-regulated (Figure 2D). Reactome pathways, including mitochondrial translation, complex Ⅰ biogenesis, and respiratory electron transport, were down-regulated, whereas biological oxidations were up-regulated (Figure 2E). The majority of down-regulated pathways enriched were therefore mitochondrial related.
Figure 2.
Enrichment Analysis of DEPs Between WT and Lmna–/– Mice Hearts
(A) Principal component analysis (PCA) of proteins in 1-month-old WT and Lmna–/– hearts. (B) PCA of proteins in 2-week-old WT and Lmna–/– hearts. (C) Venn diagram depicting differential and overlapping differentially expressed proteins (DEPs) in 1-month-old and 2-week-old WT and Lmna–/– hearts (upper); 257 DEPs in 1 month consistent with the trend of change at 2 weeks of age (lower). (D) Gene Set Enrichment Analysis showing enriched top 10 up-regulated and down-regulated Kyoto Encyclopedia of Genes and Genomes pathways. (E) Gene Set Enrichment Analysis showing enriched top 10 up-regulated and down-regulated Reactome pathways. (F) Heatmaps summarizing differentially expressed mitochondrial respiratory chain Ⅰ, Ⅲ, and Ⅳ assembly proteins. (G) Representative Western blot images of electron transport chain complex subunits (complexes I-V) in 1-month-old WT and Lmna–/– hearts. (H) Quantification of electron transport chain complex I to V expressions normalized to β-actin (n = 3). Data are presented as mean ± SD and were compared by using Student’s t-test (panel H). ∗P < 0.05, ∗∗P < 0.01. ATP = adenosine triphosphate; cAMP = cyclic adenosine monophosphate; ns = no significant difference; PPAR = peroxisome proliferator–activated receptor; other abbreviations as in Figure 1.
Protein–protein interaction networks analysis, based on the aforementioned 257 DEPs at 1 month, found that most of the core region proteins were mitochondria related, with some being associated with acetylation. This suggests that protein modification may also participate in the pathology of cardiac dysfunction (Supplemental Figure 3D). Moreover, multiple proteins related to mitochondrial respiratory chain complexes assembly and OXPHOS were consistently down-regulated in 1-month-old Lmna–/– hearts, as shown in the heatmaps (Figure 2F, Supplemental Figure 3E); this was further verified by Western blot analysis showing decreased electron transport chain complex Ⅰ, Ⅱ, Ⅲ, and Ⅴ expression (Figures 2G and 2H). Taken together, our proteomics data indicate that mitochondrial function–related proteins are down-regulated in the hearts of Lmna–/– mice, which may promote DCM.
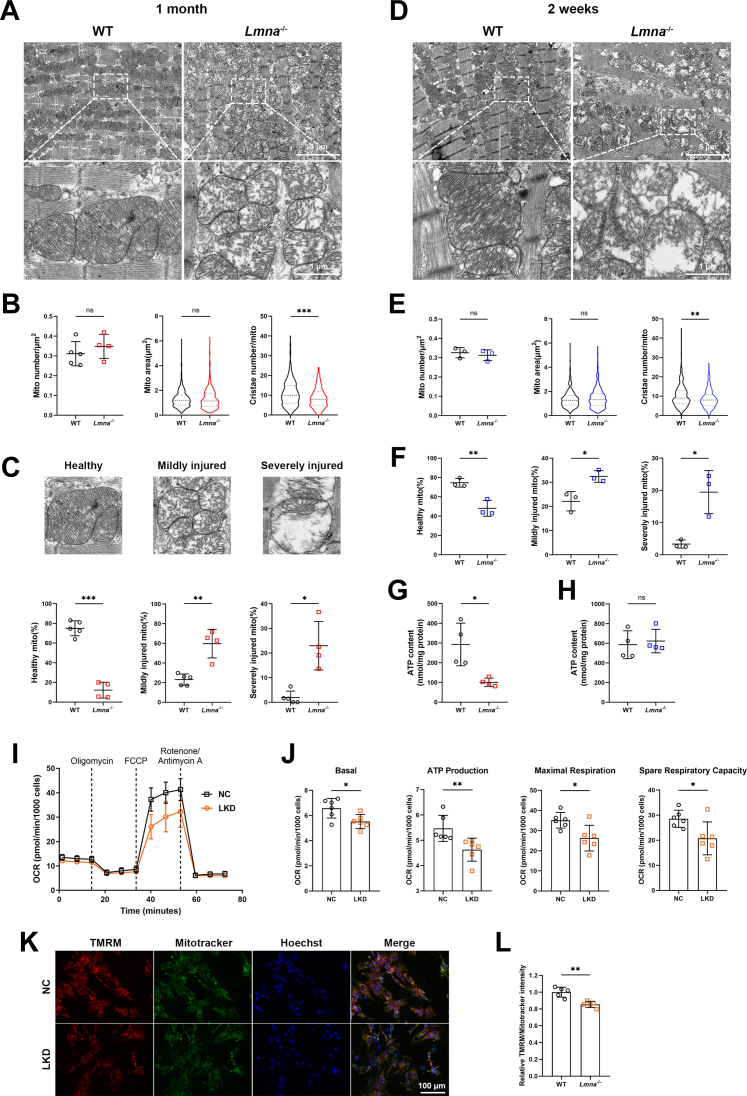
Lamin A/C Deficiency Leads to Aberrant Mitochondrial Structure and Function
To determine the mitochondrial injury caused by LMNA deficiency, the myocardial mitochondrial ultrastructure was studied using transmission electron microscopy (TEM). In line with the mass spectrometry data, mitochondrial cristae of 1-month-old Lmna–/– hearts were sparse and disorganized with decreased density (Figure 3A). Quantitative analysis revealed a marked reduction in cristae number, whereas mitochondrial area and number were not affected (Figure 3B). Furthermore, the percentage of mildly and severely injured mitochondria was much higher in Lmna–/– hearts than in WT hearts (Figure 3C). Importantly, partial mitochondrial morphology abnormalities, such as decreased cristae density and elevated injured mitochondria, were also observed in 2-week-old Lmna–/– hearts (Figures 3D to 3F), suggesting that mitochondrial impairment started in the early stage of LMNA-DCM. Coinciding with these results, the heatmap showed significantly down-regulated cristae formation proteins in 1-month-old Lmna–/– hearts (Supplemental Figure 3F). In addition to cristae structure damage, ATP content was reduced in Lmna–/– hearts at 1 month but not at 2 weeks (Figures 3G and 3H), indicating defective mitochondrial bioenergetics in the advanced stage of cardiac dysfunction caused by lamin A/C deficiency.
Figure 3.
Lamin A/C Deficiency Leads to Aberrant Mitochondrial Structure and Function
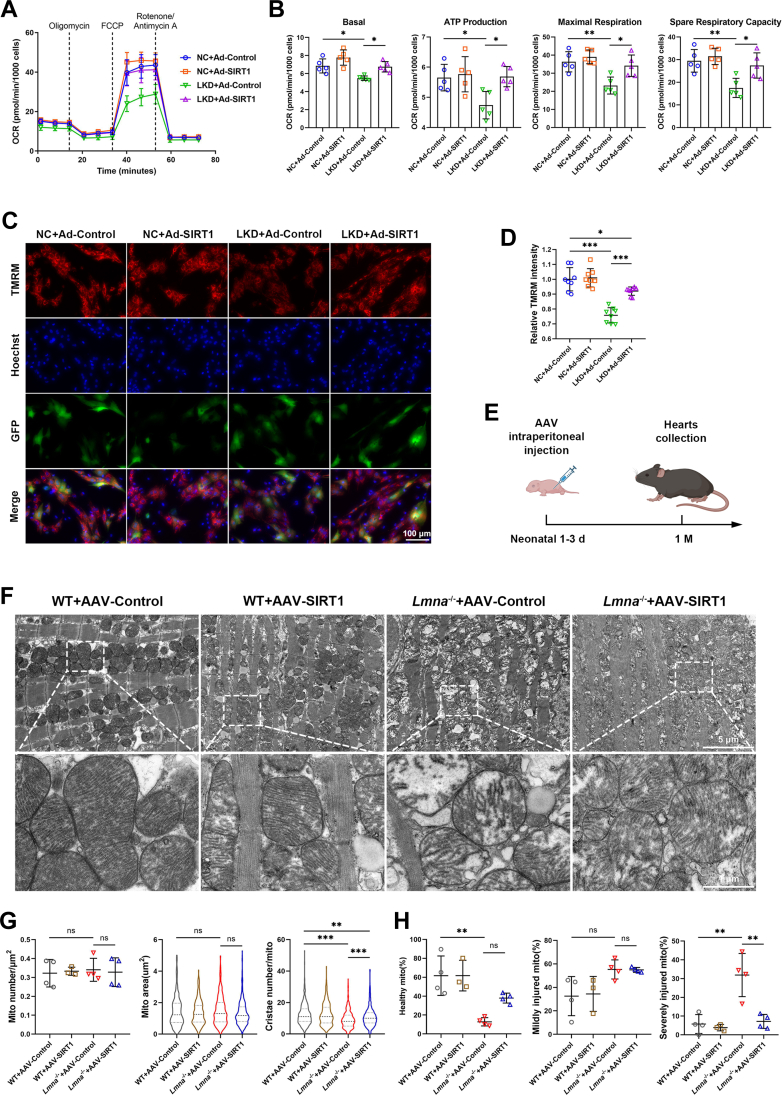
(A) Representative transmission electron microscopy images of myocardial mitochondria from 1-month-old WT and Lmna–/– mice (scale bars: 5 μm and 1 μm). (B) Quantification of mitochondrial number per square micrometers (WT, n = 5; Lmna–/–, n = 4), mitochondrial area (WT, n = 424; Lmna–/–, n = 437), and cristae number per mitochondrion (WT, n = 330; Lmna–/–, n = 354). (C) Quantification of the percentage of healthy, mildly injured, and severely injured mitochondria according to cristae density (WT, n = 5; Lmna–/–, n = 4). (D) Representative transmission electron microscopy images of myocardial mitochondria from 2-week-old WT and Lmna–/– mice. Scale bar: 5 μm and 1 μm. (E) Quantification of mitochondrial number per square micrometers (n = 3), mitochondrial area (WT, n = 238; Lmna–/–, n = 248), and cristae number per mitochondrion (WT, n = 238; Lmna–/–, n = 248). (F) Quantification of the percentage of healthy, mildly injured, and severely injured mitochondria according to cristae density (n = 3). (G and H) ATP levels in myocardial tissue of 1-month-old (G) and 2-week-old (H) WT and Lmna–/– mice (n = 4). (I) Representative image of oxygen consumption rate (OCR) in negative control (NC) and lamin A/C knockdown (LKD) neonatal rat ventricular myocytes (NRVMs). (J) Quantification of basal respiration, ATP production, maximal respiration, and spare respiratory capacity (n = 6). (K) Representative fluorescence images showing mitochondrial membrane potential of NC and LKD NRVMs stained by tetramethylrhodamine methyl ester (TMRM) and MitoTracker Green (scale bar: 100 μm). (L) Quantification of TMRM intensity (n = 8). Data are presented as mean ± SD. Data in B (mito number), C, E (mito number), F, G, J, and L were compared by using the Student’s t-test, and data in B (mito area, cristae number), E (mito area, cristae number), and H were compared by using the Mann-Whitney U test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Abbreviations as in Figures 1 and 2.
We next sought to determine whether loss of lamin A/C results in mitochondrial dysfunction in vitro. NRVMs were transfected with siRNA targeting rat Lmna, which reduced the lamin A/C protein level by 90% (Supplemental Figures 4A and 4B). The Seahorse Cell Mito Stress test showed that lamin A/C knockdown (LKD) NRVMs had reduced basal respiration, ATP production, maximal respiration, and spare respiration capacity, albeit unaffected proton leak and non-mitochondrial oxygen consumption (Figures 3I and 3J, Supplemental Figure 4C). In addition, collapsed mitochondrial membrane potential was detected in LKD NRVMs by tetramethylrhodamine methyl ester staining, which was normalized by MitoTracker signal (Figures 3K and 3L); the mitochondrial oxidative stress level as measured by MitoSOX remained unchanged (Supplemental Figures 4D and 4E). Collectively, these data indicate that LMNA impairment leads to mitochondrial structural and functional abnormalities, which preceded the DCM phenotype.
Lamin A/C Deficiency Causes Impaired Mitochondrial Biogenesis, Imbalanced Fission and Fusion, and Defective Mitophagy
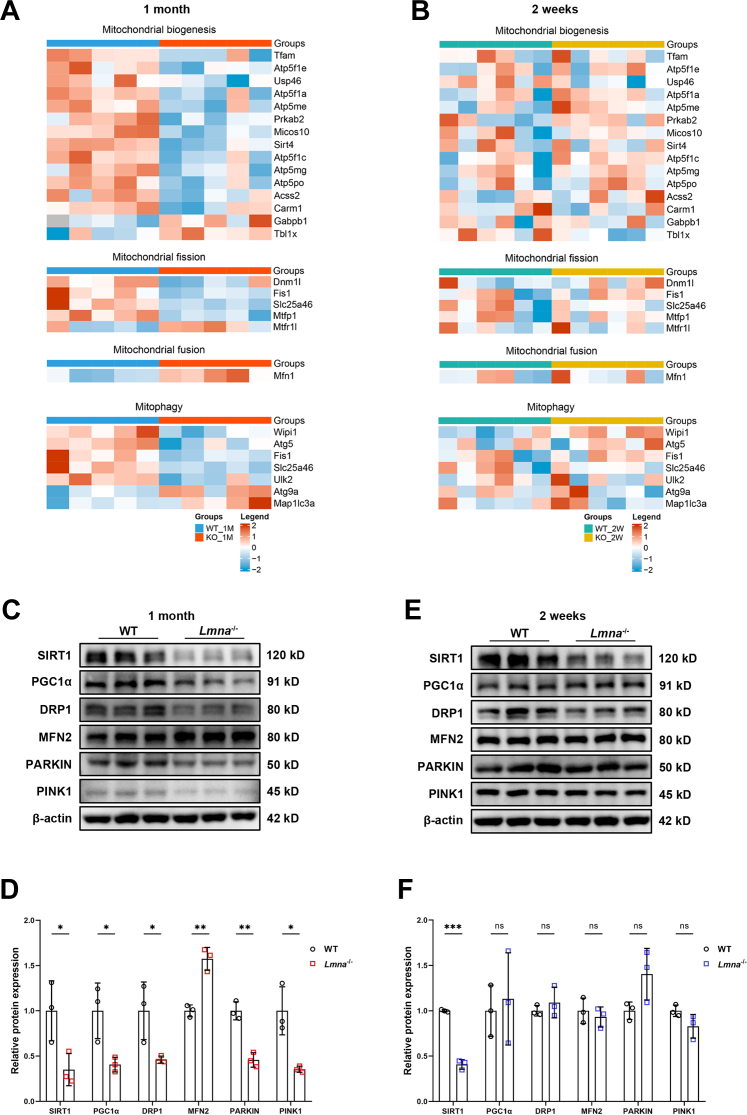
Mitochondria are semi-autonomous organelles that continuously undergo biogenesis, fission, fusion, and mitophagy to counteract stress and maintain their health and integrity. These processes are termed mitochondrial quality control (MQC).36 Abnormalities or dissonance of these biological processes culminate in defective mitochondrial bioenergetics, which are critical in various of cardiovascular diseases.23,37 We therefore investigated the key regulators of these processes in lamin A/C–deficient conditions. Heatmaps depicted those proteins related to mitochondrial biogenesis (eg, Tfam, Sirt4), fission (eg, Dnm1l, Fis1), and mitophagy (eg, Atg5) were significantly down-regulated, whereas the fusion-related protein Mfn1 was up-regulated in 1-month-old but not 2-week-old Lmna–/– hearts (Figures 4A and 4B). Consistently, the master mitochondrial biogenesis regulator SIRT1, as well as its downstream target PGC1α, was significantly down-regulated in 1-month-old Lmna–/– hearts (Figures 4C and 4D). Moreover, DRP1, which promotes fission, was also reduced, although its counterpart MFN2, which promotes fusion, was markedly up-regulated, suggesting an imbalance between mitochondrial fission and fusion. In addition, 2 essential mitophagy regulatory proteins, PARKIN and PINK1, were both markedly decreased, suggesting an impaired ability to eliminate damaged mitochondria in the end stage of LMNA-DCM. Similar changes at the mRNA levels were confirmed by Real-time Quantitative Polymerase Chain Reaction (RT-qPCR) (Supplemental Figure 5A). In contrast, among these regulators, only SIRT1 was down-regulated in 2-week-old Lmna–/– hearts (Figures 4E and 4F), suggesting that SIRT1 reduction is an early event in lamin A/C deficiency–induced DCM. However, RT-qPCR showed that SIRT1 mRNA levels were comparable between Lmna–/– and WT hearts, supporting a posttranscriptional regulatory mechanism.
Figure 4.
Lamin A/C Deficiency Causes Impaired Mitochondrial Biogenesis, Imbalanced Fission and Fusion, and Defected Mitophagy
(A and B) Heatmaps summarizing DEPs related to mitochondrial quality control (MQC) in 1-month-old (A) and 2-week-old (B) WT and Lmna–/– hearts. (C to F) Representative Western blot images of MQC-related proteins in 1-month-old (C) and 2-week-old (E) WT and Lmna–/– hearts. Quantification of MQC-related proteins expressions normalized to β-actin in 1-month-old (D) and 2-week-old (F) WT and Lmna–/– hearts (n = 3). Data are presented as mean ± SD and were compared by using Student’s t-tests (D and F). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. DRP1 = dynamin-related protein 1; MFN2 = mitofusin 2; PGC1α, peroxisome proliferator–activated receptor gamma coactivator 1-alpha; PINK1 = PTEN-induced kinase 1; SIRT1 = sirtuin 1; other abbreviations as in Figures 1 and 2.
We next studied mitochondrial morphology in MitoTracker Red–stained cardiomyocytes using OMX imaging. In LKD NRVMs, perinuclear mitochondria were increased, but intermyofibrillar mitochondria were more sparse (Supplemental Figure 5B), suggesting less efficient energy supply for sarcomere contraction. Quantification of the mitochondrial network revealed a decreased mitochondrial footprint and branched networks (Supplemental Figure 5C), which may result from impaired mitochondrial biogenesis. Together, these results show that defective mitochondrial bioenergetics in the lamin A/C–deficient heart at 1 month is associated with dysregulation in multiple MQC regulators, among which only SIRT1 reduction was found in 2-week-old Lmna–/– hearts, suggesting it as a possible mechanism for initiating DCM.
SIRT1 Overexpression Prevents Cardiac Mitochondrial Injury Caused by Lamin A/C Deficiency
SIRT1 is known to promote mitochondrial biogenesis and function, and its deletion in cardiomyocytes induces DCM.38,39 Given that SIRT1 was down-regulated in Lmna–/– hearts at 2 weeks and 1 month, we hypothesized that SIRT1 reduction contributes to the mitochondrial injury and heart failure with lamin A/C impairment. LKD NRVMs were infected with adenovirus expressing SIRT1-3×FLAG-eGFP (Ad-SIRT1) or adenovirus expressing eGFP only (Ad-Control). Seahorse analysis revealed that SIRT1 overexpression restored OXPHOS levels in LKD NRVMs, with increased basal respiration, ATP production, maximal respiration, and spare respiration capacity (Figures 5A and 5B). In addition, the reduced mitochondrial membrane potential in LKD NRVMs was also normalized by SIRT1 overexpression (Figures 5C and 5D). These results suggest that SIRT1 overexpression rescues the bioenergetics impairment in lamin A/C–deficient cardiomyocytes.
Figure 5.
Overexpression of SIRT1 Prevents Cardiac Mitochondrial Injury Caused by Lamin A/C Deficiency
(A) Representative image of mitochondrial OCR in NC and LKD NRVMs transfected with adenovirus expressing SIRT1-3×FLAG-eGFP (Ad-SIRT1) or adenovirus expressing enhanced green fluorescent protein (eGFP) only (Ad-Control). (B) Quantification of basal respiration, ATP production, maximal respiration, and spare respiratory capacity (n = 5). (C) Representative fluorescence images showing mitochondrial membrane potential of NC and LKD NRVMs transfected with Ad-SIRT1-3×FLAG-eGFP or Ad-eGFP stained by TMRM. Scale bar: 100 μm. (D) Quantification of TMRM intensity (n = 8). (E) Adeno-associated virus (AAV) vector was intraperitoneally injected in neonatal 1-3 days old WT and Lmna–/– mice. Hearts were collected at 1 month for further analysis. (F) Representative transmission electron microscopy images of myocardial mitochondria from 1-month-old WT and Lmna–/– mice pretreated with AAV-CMV-SIRT1-mNeonGreen (AAV-SIRT1) or AAV-CMV-mNeonGreen (AAV-Control). Scale bars: 5 μm and 1 μm. (G) Quantification of mitochondrial number per square micrometers (WT+AAV-Control, Lmna–/–+AAV-Control, Lmna–/–+AAV-SIRT1, n = 4; WT+AAV-SIRT1, n = 3), mitochondrial area (WT+AAV-Control, n = 318; WT+AAV-SIRT1, n = 263; Lmna–/–+AAV-Control, n = 420; Lmna–/–+AAV-SIRT1, n = 397), and cristae number per mitochondrion (WT+AAV-Control, n = 319; WT+AAV-SIRT1, n = 263; Lmna–/–+AAV-Control, n = 420; Lmna–/–+AAV-SIRT1, n = 397). (H) Quantification of the percentage of healthy, mildly injured, and severely injured mitochondria according to cristae density (WT+AAV-Control, Lmna–/–+AAV-Control, Lmna–/–+AAV-SIRT1, n = 4; WT+AAV-SIRT1, n = 3). Data are presented as mean ± SD. Data in B, D, G (mito number), and H were compared by using an analysis of variance with the Tukey post hoc test, and data in G (mito area, cristae number) were compared by using the Kruskal-Wallis test followed by the Dunn post hoc test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Abbreviations as in Figure 1, Figure 2, Figure 3, Figure 4.
We next evaluated the therapeutic effect of SIRT1 on mitochondrial injury in Lmna–/– mice. Adeno-associated virus AAV-CMV-SIRT1-mNeonGreen (AAV-SIRT1) or AAV-CMV-mNeonGreen (AAV-Control) were injected in neonatal 1 to 3 days old mice, and hearts were collected for analyses at 1 month of age (Figure 5E). Up-regulation of SIRT1 mRNA and protein was verified in AAV-SIRT1 mice (Supplemental Figures 6A to 6C), and TEM analysis indicated that the decreased mitochondrial cristae density and aberrant morphology in Lmna–/– hearts were partially restored by SIRT1 overexpression (Figure 5F). Quantification of TEM images showed increases in mitochondrial cristae number and percentage of healthy mitochondria, as well as a reduced number of severely injured mitochondria in Lmna–/– hearts with AAV-SIRT1 compared with Lmna–/– hearts with AAV-Control (Figures 5G and 5H), suggesting a key role of SIRT1 in maintaining mitochondrial health in vivo. These observations suggest that SIRT1 protects against cardiac mitochondrial injury induced by lamin A/C deficiency.
AAV-Mediated SIRT1 Overexpression Alleviates Cardiac Dysfunction and Prolongs Survival in Lmna–/– Mice
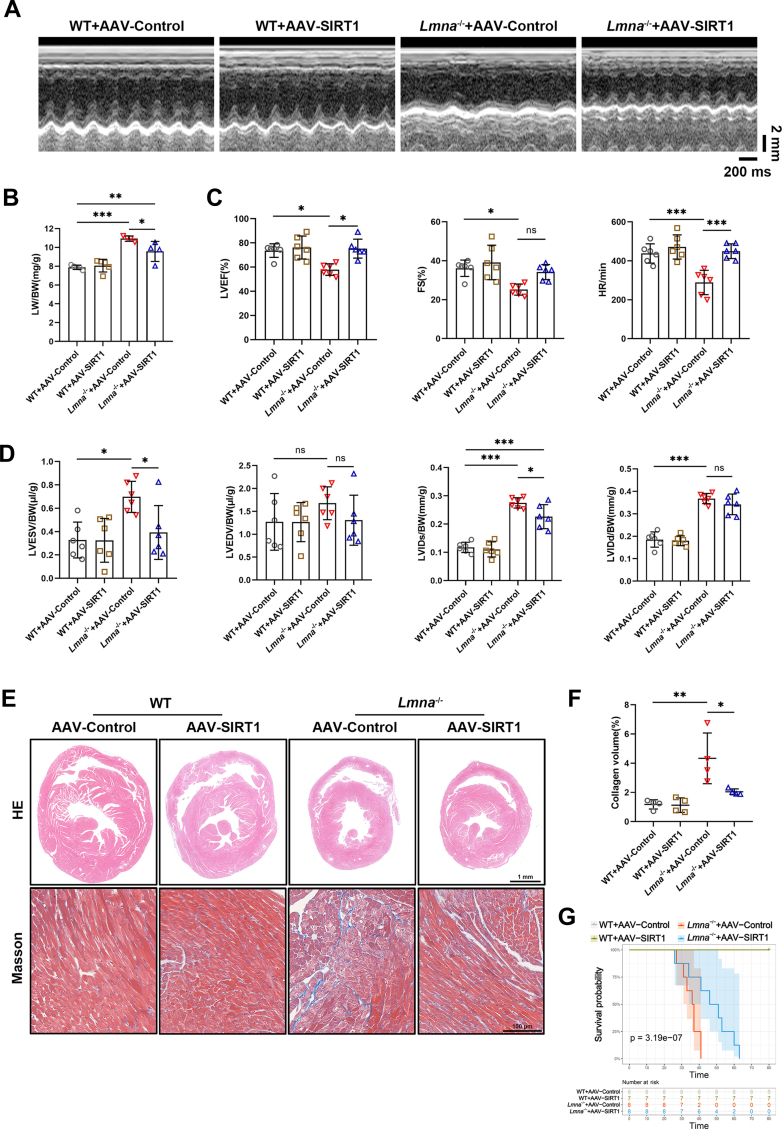
We next sought to determine the therapeutic effect of AAV-SIRT1 pretreatment on the cardiac phenotype of Lmna–/– mice. At 1 month post–AAV-SIRT1 injection, no significant improvements were found in exteriors, BW, and HW, nor in the HW/BW, of Lmna–/– mice compared with the AAV-Control group (Supplemental Figures 6D and 6E). However, increased LW/BW in 1 month-old Lmna–/– mice was mitigated by pretreatment with AAV-SIRT1 (Figure 6B). Echocardiographic analysis revealed marked increases in LVEF, FS, and heart rate in Lmna–/– mice pretreated with AAV-SIRT1 (Figures 6A and 6C). Furthermore, the abnormal LVESV/BW and left ventricular internal diameter at end-systole/BW in Lmna–/– mice were attenuated by AAV-SIRT1, whereas LVEDV/BW and left ventricular internal diameter at end-diastole/BW remained unchanged (Figure 6D).
Figure 6.
SIRT1-Based AAV Injection Alleviates Cardiac Dysfunction and Prolongs Survival of Lmna–/– Mice
(A) Representative echocardiography of WT and Lmna–/– mice pretreated with AAV-SIRT1 or AAV-Control. (B) LW/BW of WT and Lmna–/– mice pretreated with AAV-SIRT1 or AAV-Control (WT+AAV-Control, WT+AAV-SIRT1, n = 5; Lmna–/–+AAV-Control, Lmna–/–+AAV-SIRT1, n = 4). (C) Quantification of LVEF, FS, and HR (n = 6). (D) Quantification of LVESV/BW, LVEDV/BW, LVIDs/BW, and LVIDd/BW (n = 6). (E) Representative HE staining (scale bar: 1 mm) and Masson trichrome staining (scale bar: 100 μm) images of the ventricular transverse section of WT and Lmna–/– mice pretreated with AAV-SIRT1 or AAV-Control. (F) Quantification of the percentage of collagen in myocardial tissue (n = 4). (G) Kaplan-Meier curves of Lmna–/– mice pretreated with AAV-Control (median survival 36.5 days) or AAV-SIRT1 (median survival 48.5 days) (WT+AAV-Control, Lmna–/–+AAV-Control, Lmna–/–+AAV-SIRT1, n = 8; WT+AAV-SIRT1, n = 7; Breslow generalized Wilcoxon test). Data are presented as mean ± SD. Data in B, C (HR), D, and F were compared by using an analysis of variance with a Tukey post hoc test; data in C (LVEF and FS) were compared by using the Kruskal-Wallis test with the Dunn post hoc test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Abbreviations as in Figures 1, 3, 4, and 5.
Histological analysis revealed mitigated ventricular dilation and reduced cardiac fibrosis in Lmna–/– mice with AAV-SIRT1, although the heart size was not altered (Figures 6E and 6F). Moreover, AAV-SIRT1 administration did not affect basal heart function or geometry in WT mice, suggesting no obvious side effects of AAV-based SIRT1 gene therapy. Although Lmna–/– mice with AAV-Control lived a median survival time of 36.5 days, Lmna–/– mice with AAV-SIRT1 lived to a median of 48.5 days (Figure 6G), which is a significant increase in longevity. These findings suggest that AAV-mediated SIRT1 overexpression corrects the cardiac dysfunction in Lmna–/– mice, implicating a therapeutic potential for cardiac laminopathy.
Lamin A/C Maintains Mitochondrial Bioenergetics Through the SIRT1-PARKIN Axis
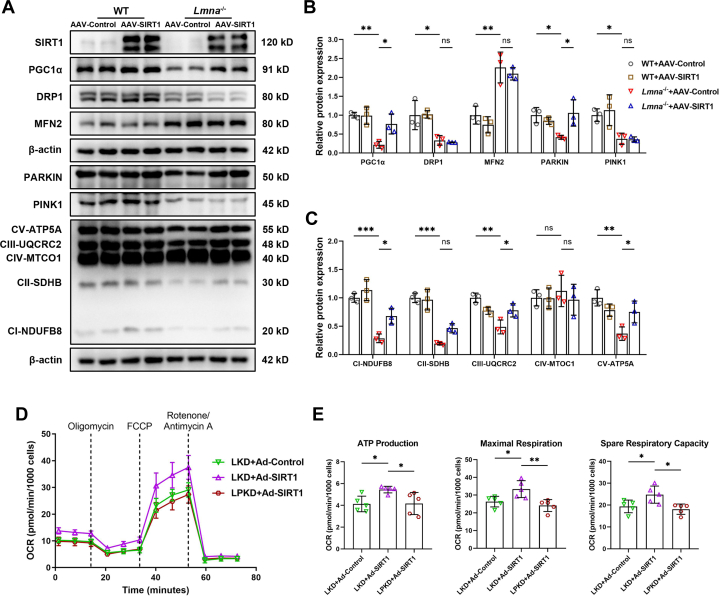
To decipher the underlying mechanism that SIRT1 protects mitochondrial bioenergetics in lamin A/C–deficient hearts, we tested mitochondrial homeostasis–related protein expressions in 1-month-old Lmna–/– hearts with AAV-SIRT1 administration. As shown in Figures 7A to 7C, the SIRT1 target PGC1α and the mitophagy mediator PARKIN, as well as complexes Ⅰ, Ⅱ, Ⅲ, and Ⅴ, were significantly up-regulated in Lmna–/– mice with AAV-SIRT1 compared with AAV-Control. However, DRP1, MFN2, and PINK1 levels were unaltered by AAV-SIRT1 (Figures 7A and 7B). These results suggest that SIRT1 maintains mitochondrial energy metabolism through PGC1α and PARKIN.
Figure 7.
Lamin A/C Maintain Mitochondrial Bioenergetics Through SIRT1-PARKIN Axis
(A) Representative Western blot images of MQC-related proteins and electron transport chain (ETC) complex subunits (complex I-V) in WT and Lmna–/– mice pretreated with AAV-SIRT1 or AAV-Control. (B and C) Quantification of MQC-related proteins (B) and complex I to V (C) expressions normalized to β-actin (n = 3). (D) Representative image of OCR in LKD NRVMs transfected with Ad-Control or Ad-SIRT1, and lamin A/C and PARKIN knockdown (LPKD) NRVMs transfected with Ad-SIRT1. (E) Quantification of ATP production, maximal respiration, and spare respiratory capacity (n = 5). Data are presented as mean ± SD and were compared by using analysis of variance with a Tukey post hoc test (B, C, and E). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Abbreviations as in Figure 1, Figure 2, Figure 3, Figure 4, Figure 5.
Considering the known protective role of SIRT1-PGC1α in energy metabolism,40 we next investigated if PARKIN is necessary for the advantageous role of SIRT1 on mitochondrial bioenergetics. PARKIN was knocked down in LKD NRVMs using siRNA (knockdown of both Lmna and Parkin). As expected, PARKIN knockdown completely abrogated the promotive effect of SIRT1 on oxidative respiratory capacity in lamin A/C deficiency (Figures 7D and 7E), suggesting that lamin A/C modulates mitochondrial function through, at least partially, the SIRT1-PARKIN axis.
Discussion
DCM patients with LMNA mutations frequently manifest with more severe heart failure, earlier malignant arrhythmias, and sudden death, with nearly 20% requiring heart transplantation, yet the pathogenesis of LMNA-DCM is not fully understood.41,42 The present study showed that mitochondrial injury is an early event in Lmna–/– mice hearts, as evidenced by the progressively down-regulated pathways related to mitochondrial translation, assembly of respiratory chain complexes, and OXPHOS, along with premature compromised cristae structure and number, which may be secondary to down-regulation of SIRT1. Overexpression of SIRT1 ameliorates cardiac mitochondrial dysfunction both in vitro and in vivo, as well as the DCM phenotype in Lmna–/– mice. Mechanistically, PARKIN knockdown nullifies the advantageous role of SIRT1 on mitochondrial bioenergetics, suggesting that lamin A/C protects mitochondrial bioenergetics at least partially via the SIRT1-PARKIN axis. This study suggests that targeting the SIRT1 signaling pathway is a potential novel therapeutic avenue for DCM with LMNA mutations.
In the present study, we generated Lmna–/– mice using CRISPR/Cas9 technology and found that 1-month-old mice exhibited cardiac dysfunction, ventricular dilation, and fibrosis, but 2-week-old mice still showed normal cardiac function. This study was the first to perform myocardial tissue proteomics of Lmna–/– hearts at ages 2 weeks and 1 month to unmask alterations in signal transduction pathways at the pre-DCM and DCM-onset stages. Our enrichment analysis from Kyoto Encyclopedia of Genes and Genomes and Reactome revealed concordant down-regulation of mitochondria-related pathways such as OXPHOS, mitochondrial translation, and respiratory electron transport, suggesting that mitochondrial dysfunction plays a contributing role in the development of DCM caused by LMNA deletion. Consistently, early-stage severely injured mitochondria with cristae loss were observed by using TEM, along with decreased cristae formation proteins. In addition, our proteomics data also revealed other dysregulated biological processes, such as protein processing, closely related to the function of the endoplasmic reticulum,43,44 which may accentuate cardiac dysfunction of Lmna–/– mice. Furthermore, a number of identified DEPs were pertinent to acetylation, a posttranslational modification that can be modulated by SIRT1.45 In addition, extracellular flux analysis revealed defective ATP production and maximal and spare respiration capacity in LKD cardiomyocytes, suggesting an inability to meet cell energetic demands under certain stressful conditions when lamin A/C is lost.
Mitochondrial biogenesis, fission, fusion, and mitophagy are crucial for mitochondrial self-renewal and quality control. Impairment of MQC mechanisms can aggravate mitochondrial dysfunction.36,46 Our study found reduced mitochondrial biogenesis regulators (SIRT1, PGC1α, and TFAM), fission-related proteins (DRP1 and FIS1), and down-regulated mitophagy markers (PARKIN and PINK1), but elevated fusion-related proteins (MFN1 and MFN2), in the myocardium of 1-month-old Lmna–/– mice, indicating MQC disorders. Notably, only SIRT1 reduction was found in 2 week-old Lmna–/– hearts, suggesting an early culprit role of mitochondrial biogenesis disruption in LMNA-DCM. Mitochondrial biogenesis is an intricate nuclear-mitochondrial process, which involves a symmetrical fission that generates 2 healthy mitochondria to increase mitochondrial mass and quantity.47,48 This process is triggered by SIRT1 and its deacetylation of PGC1α.20,49 SIRT1-knockout mice develop DCM, accompanied by a decrease in the volume of cardiomyocytes, and abnormal mitochondrial morphology and function.39 Recent studies have found that lamin A/C deficiency leads to bioenergetics impairment in mouse embryonic fibroblasts via attenuating PGC1α and nicotinamide phosphoribosyltransferase/NAD+ signaling,50 highlighting an obligatory role of mitochondrial biogenesis in LMNA protection of mitochondrial function, which is consistent with our results. Mitochondrial fission and fusion are mutually antagonistic but related processes that are essential to ensure optimal mitochondrial function under physiological and pathologic contexts.51 Although excessively activated fission and down-regulated fusion were observed in some cardiovascular diseases such as cardiac hypertrophy, ischemia-reperfusion injury, or heart failure,52, 53, 54 our data indicated inhibited fission and enhanced fusion, suggesting that distinct regulation mechanisms exist in Lmna–/– hearts. Of note, deletion or mutation of DRP1 in mice hearts also leads to heart failure and premature death, concomitant with mitophagy defects.55,56 Overexpression of MFN2 can bring about mitochondrial perinuclear aggregation, a phenomenon before apoptotic cell death,57 which was also observed from our fluorescence imaging of LKD NRVMs. Therefore, whether this imbalance of mitochondrial fission and fusion in the Lmna–/– hearts exacerbates DCM or simply acts as a protective compensatory mechanism under cardiac stress warrants further investigation.
Given that SIRT1 is down-regulated before the emergence of evident cardiac dysfunction in Lmna–/– mice, as well as its regulatory role in energy metabolism,21 we investigated whether SIRT1 overexpression can reverse mitochondrial damage and the DCM phenotype in Lmna–/– mice. As expected, SIRT1 alleviates mitochondrial structural and functional injury, mitigates cardiac systolic dysfunction and fibrosis, and extends the lifespan of Lmna–/– mice. Previous findings have shown that lamin A activates SIRT1 deacetylase, and Lmna–/– mouse embryonic fibroblasts exhibit decreased SIRT1 deacetylase activity.25,50 Thus, SIRT1 activators may be an option for the treatment of LMNA-DCM. The AAV9 vector requires only a single injection, and the in vivo transgene expressions can be detected after 1 year, which has been successfully used in LMNA-associated cardiomyopathy mouse models.12,58 Furthermore, AAV-based gene therapy has been shown to be safe for clinical application, including in patients with cardiovascular diseases.59,60 SIRT1-targeted therapy is clinically relevant. First, there is clinical evidence that SIRT1 is down-regulated in myocardial samples from heart failure patients with ischemic cardiomyopathy and DCM, as well as in patients with diabetes. In addition, reduced SIRT1 expression in the myocardium of patients with diabetes is associated with an increase in intramyocardial triacylglycerol content and up-regulation of peroxisome proliferator–activated receptor gamma signaling.61,62 Second, SIRT1 has been proposed as a downstream mechanism of cardioprotective effects of sodium-glucose cotransporter 2 inhibitors, which have been shown in large-scale clinical trials to reduce heart failure hospitalizations and cardiovascular death.63 Finally, SIRT1 is highly dependent on the bioavailability of NAD+. Boosting NAD+ has been proven to be a promising therapeutic avenue against cardiomyopathy and heart failure in several mice models and human studies.64, 65, 66, 67 Together, our results support prior research and have favorable clinical transformation prospects.
Mechanistically, we found that PARKIN knockdown nullified the beneficial effects of SIRT1 on energy metabolism in LKD NRVMs, suggesting PARKIN is necessary for mitochondrial bioenergetics downstream of SIRT1 in lamin A/C–deficient conditions. PARKIN is an E3 ubiquitin protein ligase required for mitophagy activation. Neonatal mice with PARKIN knockout develop perinatal cardiomyopathy in concert with premature death due to a disruption of fetal mitochondrial mitophagic removal.68 SIRT1-PARKIN–mediated mitophagy has been shown to be pivotal in cardiac senescence and ischemia/reperfusion injury.69,70 Our results showed that the SIRT1-PARKIN axis improves mitochondrial oxidative respiration capacity in lamin A/C–deficient cardiomyocytes; however, detailed evaluation of PARKIN-mediated mitophagy and its alternation following the progression of cardiac dysfunction in Lmna–/– hearts requires further studies.
Study Limitations
Although our findings suggest a new strategy for treating LMNA-DCM, there are several limitations. First, the Lmna–/– mice model used in this study was constructed by global knockout of the Lmna gene. Although these mice faithfully exhibit the DCM phenotype, they do not recapitulate all the genotypes of DCM patients with LMNA mutations, some of which have gain-of-function or are heterozygous. In addition, the high mortality of Lmna–/– mice is likely of cardiac origins but may also involve other tissue and organ failure. Likewise, although the endpoints are primarily cardiac, it is noteworthy that the benefits of SIRT1 could potentially be attributed to the off-targeting effects of AAV9, including vascular endothelium dysfunction restoration,71 skeletal muscle and respiratory function improvement,72,73 and adipose tissue metabolic regulation.74 Cardiomyocyte-specific knockout mice would be a more appropriate model for a cardiac-specific signaling pathway study. Second, arrhythmias are another mechanism of premature death in Lmna–/– mice, which were not evaluated in this study. It has been shown that SIRT1 deacetylates Nav1.5 and SERCA2a, regulating their activities.75,76 Hence, the beneficial effects of SIRT1 overexpression may also be related to changes in cardiac electrical activity. Third, it remains unclear how lamin A/C loss induces the down-regulation of SIRT1. Our preliminary data revealed an unaltered SIRT1 mRNA level after LMNA deletion, suggesting that it is unlikely a transcriptional regulation. Although previous findings suggested that lamin A interacts with SIRT1 in mouse embryonic fibroblasts and HEK293 cells,25 whether this interaction occurs in cardiomyocytes and, if so, how it contributes to SIRT1 degradation warrant further studies.
Conclusions
We found that Lmna–/– mice hearts exhibit mitochondrial structural and functional damage, as well as MQC disorders, including mitochondrial biogenesis damage, fission and fusion imbalance, and mitophagy defects. Overexpression of SIRT1 assuages the cardiac mitochondrial anomaly caused by lamin A/C deficiency and DCM phenotype in Lmna–/– mice. Mechanistically, this study elucidates that LMNA protects mitochondrial bioenergetics through the SIRT1-PARKIN axis, denoting novel therapeutic targets for LMNA-DCM.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: This study identified that mitochondrial injury and SIRT1 down-regulation are early events in Lmna–/– mice hearts. Overexpression of SIRT1 alleviates myocardial mitochondrial dysfunction and DCM caused by lamin A/C deficiency. LMNA regulates mitochondrial bioenergetics through the SIRT1-PARKIN axis.
TRANSLATIONAL OUTLOOK: Pharmacologic activation of the SIRT1 signaling pathway may present a novel therapeutic avenue for treating LMNA mutation-associated DCM.
Funding Support and Author Disclosures
This work was supported by the National Natural Science Foundation of China (81570234 and 81770124), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (RC20210190), and the National Research Center for Translational Medicine Shanghai Open Project (TMSZ-2020-201 and TMSK-2021-508). The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank Bi Ke, Ultrasound Department of Shanghai Pulmonary Hospital Affiliated to Tongji University, for the support in mouse echocardiographic examinations.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
The data underlying this paper are all available within the paper and in the Supplemental Appendix. The proteome data generated can be viewed in integrated proteome resource iProX (http://www.iprox.org) with ID IPX0006466000. The normalized data file is provided as Supplemental Table 1.
Contributor Information
Tong Yin, Email: yt10961@rjh.com.cn.
Yucai Xie, Email: xyc10530@rjh.com.cn.
Appendix
References
- 1.Lin F., Worman H.J. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 2.Worman H.J. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weintraub R.G., Semsarian C., Macdonald P. Dilated cardiomyopathy. Lancet. 2017;390:400–414. doi: 10.1016/S0140-6736(16)31713-5. [DOI] [PubMed] [Google Scholar]
- 4.Glikson M., Nielsen J.C., Kronborg M.B., et al. 2021 ESC guidelines on cardiac pacing and cardiac resynchronization therapy. Eur Heart J. 2021;42:3427–3520. doi: 10.1093/eurheartj/ehab364. [DOI] [PubMed] [Google Scholar]
- 5.McNally E.M., Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muchir A., Pavlidis P., Decostre V., et al. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J Clin Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu W., Muchir A., Shan J., Bonne G., Worman H.J. Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation. 2011;123:53–61. doi: 10.1161/CIRCULATIONAHA.110.970673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muchir A., Wu W., Choi J.C., et al. Abnormal p38alpha mitogen-activated protein kinase signaling in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum Mol Genet. 2012;21:4325–4333. doi: 10.1093/hmg/dds265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J.C., Muchir A., Wu W., et al. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos F.J., Chen S.C., Garelick M.G., et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho C.Y., Jaalouk D.E., Vartiainen M.K., Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auguste G., Gurha P., Lombardi R., Coarfa C., Willerson J.T., Marian A.J. Suppression of activated FOXO transcription factors in the heart prolongs survival in a mouse model of laminopathies. Circ Res. 2018;122:678–692. doi: 10.1161/CIRCRESAHA.117.312052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S.N., Lombardi R., Karmouch J., et al. DNA damage response/TP53 pathway is activated and contributes to the pathogenesis of dilated cardiomyopathy associated with LMNA (Lamin A/C) mutations. Circ Res. 2019;124:856–873. doi: 10.1161/CIRCRESAHA.118.314238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J., Termglinchan V., Diecke S., et al. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 2019;572:335–340. doi: 10.1038/s41586-019-1406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao Z., Koh W., Ni Y., et al. RNA sequence analyses throughout the course of mouse cardiac laminopathy identify differentially expressed genes for cell cycle control and mitochondrial function. Sci Rep. 2020;10:6632. doi: 10.1038/s41598-020-63563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ignatieva E.V., Ivanova O.A., Komarova M.Y., et al. LMNA mutations G232E and R482L cause dysregulation of skeletal muscle differentiation, bioenergetics, and metabolic gene expression profile. Genes (Basel) 2020;11 doi: 10.3390/genes11091057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera-Torres J., Acin-Perez R., Cabezas-Sanchez P., et al. Identification of mitochondrial dysfunction in Hutchinson-Gilford progeria syndrome through use of stable isotope labeling with amino acids in cell culture. J Proteomics. 2013;91:466–477. doi: 10.1016/j.jprot.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Xu X., Wang D., Zheng C., et al. Progerin accumulation in nucleus pulposus cells impairs mitochondrial function and induces intervertebral disc degeneration and therapeutic effects of sulforaphane. Theranostics. 2019;9:2252–2267. doi: 10.7150/thno.30658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karbasforooshan H., Karimi G. The role of SIRT1 in diabetic cardiomyopathy. Biomed Pharmacother. 2017;90:386–392. doi: 10.1016/j.biopha.2017.03.056. [DOI] [PubMed] [Google Scholar]
- 20.Price N.L., Gomes A.P., Ling A.J., et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Q., Su H., Qi B., et al. A SIRT1 activator, ginsenoside Rc, promotes energy metabolism in cardiomyocytes and neurons. J Am Chem Soc. 2021;143:1416–1427. doi: 10.1021/jacs.0c10836. [DOI] [PubMed] [Google Scholar]
- 22.Qiao H., Ren H., Du H., Zhang M., Xiong X., Lv R. Liraglutide repairs the infarcted heart: the role of the SIRT1/Parkin/mitophagy pathway. Mol Med Rep. 2018;17:3722–3734. doi: 10.3892/mmr.2018.8371. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Picca A., Mankowski R.T., Burman J.L., et al. Mitochondrial quality control mechanisms as molecular targets in cardiac ageing. Nat Rev Cardiol. 2018;15:543–554. doi: 10.1038/s41569-018-0059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao J., Wang J., Xu Y., et al. CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy. 2022;18:1879–1897. doi: 10.1080/15548627.2021.2007027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B., Ghosh S., Yang X., et al. Resveratrol rescues SIRT1-dependent adult stem cell decline and alleviates progeroid features in laminopathy-based progeria. Cell Metab. 2012;16:738–750. doi: 10.1016/j.cmet.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh S., Liu B., Wang Y., Hao Q., Zhou Z. Lamin A is an endogenous SIRT6 activator and promotes SIRT6-mediated DNA repair. Cell Rep. 2015;13:1396–1406. doi: 10.1016/j.celrep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Sun S., Qin W., Tang X., et al. Vascular endothelium-targeted Sirt7 gene therapy rejuvenates blood vessels and extends life span in a Hutchinson-Gilford progeria model. Sci Adv. 2020;6 doi: 10.1126/sciadv.aay5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam J., Katti P., Biete M., et al. A universal approach to analyzing transmission electron microscopy with ImageJ. Cells. 2021;10 doi: 10.3390/cells10092177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranjbarvaziri S., Kooiker K.B., Ellenberger M., et al. Altered cardiac energetics and mitochondrial dysfunction in hypertrophic cardiomyopathy. Circulation. 2021;144:1714–1731. doi: 10.1161/CIRCULATIONAHA.121.053575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valente A.J., Maddalena L.A., Robb E.L., Moradi F., Stuart J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Du Z., Zhu T., Lin M., et al. A novel mutation in human EMD gene and mitochondrial dysfunction in emerin knockdown cardiomyocytes. J Cell Mol Med. 2022;26:5054–5066. doi: 10.1111/jcmm.17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turesson I., Velez R., Kristinsson S.Y., Landgren O. Patterns of improved survival in patients with multiple myeloma in the twenty-first century: a population-based study. J Clin Oncol. 2010;28:830–834. doi: 10.1200/JCO.2009.25.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasi Tandefelt D., Boormans J.L., van der Korput H.A., Jenster G.W., Trapman J. A 36-gene signature predicts clinical progression in a subgroup of ERG-positive prostate cancers. Eur Urol. 2013;64:941–950. doi: 10.1016/j.eururo.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan T., Escalante-Alcalde D., Bhatt H., et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolova V., Leimena C., McMahon A.C., et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L., Wang L., Du Y., Zhang Y., Ren J. Mitochondrial quality control mechanisms as therapeutic targets in doxorubicin-induced cardiotoxicity. Trends Pharmacol Sci. 2023;44:34–49. doi: 10.1016/j.tips.2022.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H., Ren J., Toan S., Mui D. Role of mitochondrial quality surveillance in myocardial infarction: from bench to bedside. Ageing Res Rev. 2021;66 doi: 10.1016/j.arr.2020.101250. [DOI] [PubMed] [Google Scholar]
- 38.Ma S., Feng J., Zhang R., et al. SIRT1 activation by resveratrol alleviates cardiac dysfunction via mitochondrial regulation in diabetic cardiomyopathy mice. Oxid Med Cell Longev. 2017;2017 doi: 10.1155/2017/4602715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Planavila A., Dominguez E., Navarro M., et al. Dilated cardiomyopathy and mitochondrial dysfunction in Sirt1-deficient mice: a role for Sirt1-Mef2 in adult heart. J Mol Cell Cardiol. 2012;53:521–531. doi: 10.1016/j.yjmcc.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 40.Packer M. Longevity genes, cardiac ageing, and the pathogenesis of cardiomyopathy: implications for understanding the effects of current and future treatments for heart failure. Eur Heart J. 2020;41:3856–3861. doi: 10.1093/eurheartj/ehaa360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiuchi S., Makiyama T., Aiba T., et al. Gene-based risk stratification for cardiac disorders in LMNA mutation carriers. Circ Cardiovasc Genet. 2017;10 doi: 10.1161/CIRCGENETICS.116.001603. [DOI] [PubMed] [Google Scholar]
- 42.Hasselberg N.E., Haland T.F., Saberniak J., et al. Lamin A/C cardiomyopathy: young onset, high penetrance, and frequent need for heart transplantation. Eur Heart J. 2018;39:853–860. doi: 10.1093/eurheartj/ehx596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopaschuk G.D., Karwi Q.G., Tian R., Wende A.R., Abel E.D. Cardiac energy metabolism in heart failure. Circ Res. 2021;128:1487–1513. doi: 10.1161/CIRCRESAHA.121.318241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren J., Bi Y., Sowers J.R., Hetz C., Zhang Y. Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat Rev Cardiol. 2021;18:499–521. doi: 10.1038/s41569-021-00511-w. [DOI] [PubMed] [Google Scholar]
- 45.Li P., Ge J., Li H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat Rev Cardiol. 2020;17:96–115. doi: 10.1038/s41569-019-0235-9. [DOI] [PubMed] [Google Scholar]
- 46.Bonora M., Wieckowski M.R., Sinclair D.A., Kroemer G., Pinton P., Galluzzi L. Targeting mitochondria for cardiovascular disorders: therapeutic potential and obstacles. Nat Rev Cardiol. 2019;16:33–55. doi: 10.1038/s41569-018-0074-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertero E., Maack C. Metabolic remodelling in heart failure. Nat Rev Cardiol. 2018;15:457–470. doi: 10.1038/s41569-018-0044-6. [DOI] [PubMed] [Google Scholar]
- 48.Pfanner N., Warscheid B., Wiedemann N. Mitochondrial proteins: from biogenesis to functional networks. Nat Rev Mol Cell Biol. 2019;20:267–284. doi: 10.1038/s41580-018-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventura-Clapier R., Garnier A., Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 50.Maynard S., Hall A., Galanos P., et al. Lamin A/C impairments cause mitochondrial dysfunction by attenuating PGC1alpha and the NAMPT-NAD+ pathway. Nucleic Acids Res. 2022;50:9948–9965. doi: 10.1093/nar/gkac741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorn GW., 2nd Evolving concepts of mitochondrial dynamics. Annu Rev Physiol. 2019;81:1–17. doi: 10.1146/annurev-physiol-020518-114358. [DOI] [PubMed] [Google Scholar]
- 52.Fang L., Moore X.L., Gao X.M., Dart A.M., Lim Y.L., Du X.J. Down-regulation of mitofusin-2 expression in cardiac hypertrophy in vitro and in vivo. Life Sci. 2007;80:2154–2160. doi: 10.1016/j.lfs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Disatnik M.H., Ferreira J.C., Campos J.C., et al. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J Am Heart Assoc. 2013;2 doi: 10.1161/JAHA.113.000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabbah H.N., Gupta R.C., Singh-Gupta V., Zhang K., Lanfear D.E. Abnormalities of mitochondrial dynamics in the failing heart: normalization following long-term therapy with elamipretide. Cardiovasc Drugs Ther. 2018;32:319–328. doi: 10.1007/s10557-018-6805-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ikeda Y., Shirakabe A., Maejima Y., et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res. 2015;116:264–278. doi: 10.1161/CIRCRESAHA.116.303356. [DOI] [PubMed] [Google Scholar]
- 56.Song M., Mihara K., Chen Y., Scorrano L., Dorn G.W., 2nd Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–286. doi: 10.1016/j.cmet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang P., Yu T., Yoon Y. Mitochondrial clustering induced by overexpression of the mitochondrial fusion protein Mfn2 causes mitochondrial dysfunction and cell death. Eur J Cell Biol. 2007;86:289–302. doi: 10.1016/j.ejcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Chai R.J., Werner H., Li P.Y., et al. Disrupting the LINC complex by AAV mediated gene transduction prevents progression of lamin induced cardiomyopathy. Nat Commun. 2021;12:4722. doi: 10.1038/s41467-021-24849-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 60.Ishikawa K., Weber T., Hajjar R.J. Human cardiac gene therapy. Circ Res. 2018;123:601–613. doi: 10.1161/CIRCRESAHA.118.311587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Svagusa T., Sikiric S., Milavic M., et al. Heart failure in patients is associated with downregulation of mitochondrial quality control genes. Eur J Clin Invest. 2023;53 doi: 10.1111/eci.14054. [DOI] [PubMed] [Google Scholar]
- 62.Costantino S., Mengozzi A., Velagapudi S., et al. Treatment with recombinant Sirt1 rewires the cardiac lipidome and rescues diabetes-related metabolic cardiomyopathy. Cardiovasc Diabetol. 2023;22:312. doi: 10.1186/s12933-023-02057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Packer M. Cardioprotective effects of sirtuin-1 and its downstream effectors: potential role in mediating the heart failure benefits of SGLT2 (sodium-glucose cotransporter 2) inhibitors. Circ Heart Fail. 2020;13 doi: 10.1161/CIRCHEARTFAILURE.120.007197. [DOI] [PubMed] [Google Scholar]
- 64.Walker M.A., Tian R. Raising NAD in heart failure: time to translate? Circulation. 2018;137:2274–2277. doi: 10.1161/CIRCULATIONAHA.117.032626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diguet N., Trammell S.A.J., Tannous C., et al. Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation. 2018;137:2256–2273. doi: 10.1161/CIRCULATIONAHA.116.026099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker M.A., Chen H., Yadav A., et al. Raising NAD(+) level stimulates short-chain dehydrogenase/reductase proteins to alleviate heart failure independent of mitochondrial protein deacetylation. Circulation. 2023;148:2038–2057. doi: 10.1161/CIRCULATIONAHA.123.066039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abdellatif M., Sedej S., Kroemer G. NAD(+) metabolism in cardiac health, aging, and disease. Circulation. 2021;144:1795–1817. doi: 10.1161/CIRCULATIONAHA.121.056589. [DOI] [PubMed] [Google Scholar]
- 68.Gong G., Song M., Csordas G., Kelly D.P., Matkovich S.J., Dorn G.W., 2nd Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong Y.X., Wu W.Y., Song F., Wu C., Li G.R., Wang Y. Cardiac senescence is alleviated by the natural flavone acacetin via enhancing mitophagy. Aging (Albany NY) 2021;13:16381–16403. doi: 10.18632/aging.203163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng M., Bai Y., Sun X., et al. Resveratrol reestablishes mitochondrial quality control in myocardial ischemia/reperfusion injury through Sirt1/Sirt3-Mfn2-Parkin-PGC-1alpha pathway. Molecules. 2022;27 doi: 10.3390/molecules27175545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mengozzi A., Costantino S., Paneni F., et al. Targeting SIRT1 rescues age- and obesity-induced microvascular dysfunction in ex vivo human vessels. Circ Res. 2022;131:476–491. doi: 10.1161/CIRCRESAHA.122.320888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myers M.J., Shepherd D.L., Durr A.J., et al. The role of SIRT1 in skeletal muscle function and repair of older mice. J Cachexia Sarcopenia Muscle. 2019;10:929–949. doi: 10.1002/jcsm.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y., Zhang F., Ding J. As a modulator, multitasking roles of SIRT1 in respiratory diseases. Immune Netw. 2022;22:e21. doi: 10.4110/in.2022.22.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiang L., Wang L., Kon N., et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vikram A., Lewarchik C.M., Yoon J.Y., et al. Sirtuin 1 regulates cardiac electrical activity by deacetylating the cardiac sodium channel. Nat Med. 2017;23:361–367. doi: 10.1038/nm.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gorski P.A., Jang S.P., Jeong D., et al. Role of SIRT1 in modulating acetylation of the sarco-endoplasmic reticulum Ca(2+)-ATPase in heart failure. Circ Res. 2019;124:e63–e80. doi: 10.1161/CIRCRESAHA.118.313865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.