Fig. 10.
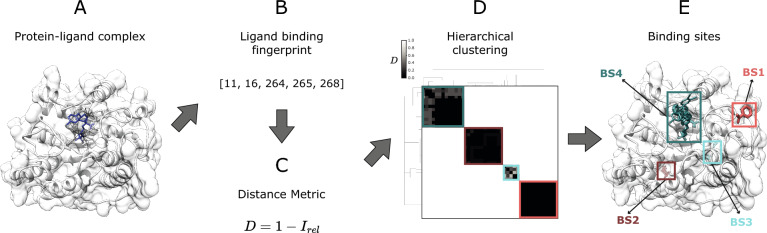
LIGYSIS ligand binding site definition algorithm. For a given protein–ligand interaction complex (A), a ligand binding fingerprint (B) is obtained as the set of unique UniProt sequence residue numbers interacting with the ligand as defined by pdbe-arpeggio; C fingerprints from different ligands binding to the same protein can be compared and a distance calculated; D this distance can be used to perform hierarchical clustering, which groups the different ligands in distinct clusters binding to the same region of the protein or binding site (E)