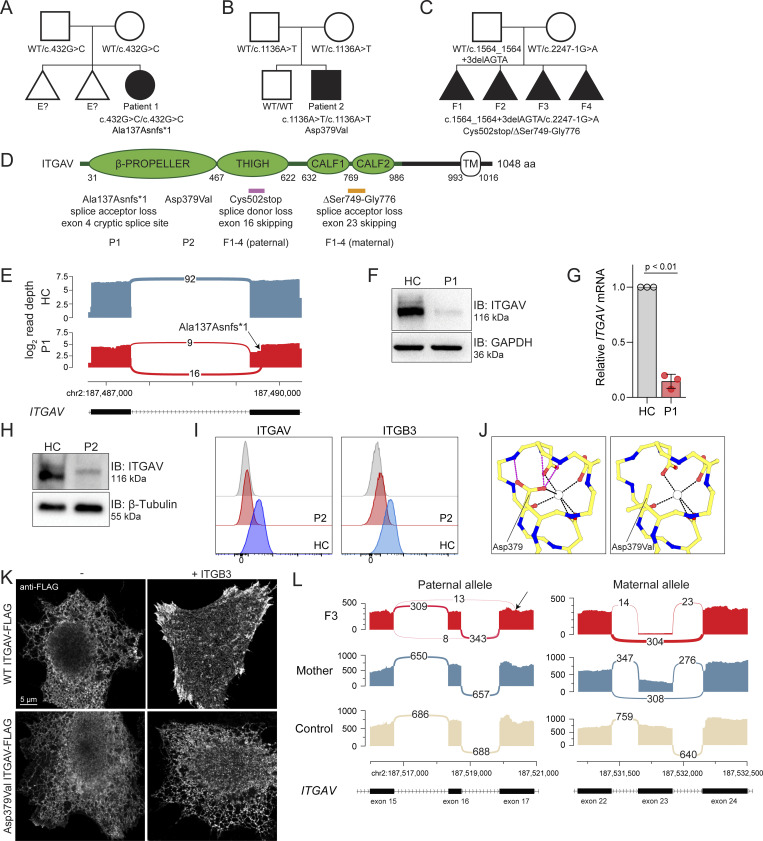
Figure 2.
Genetic and functional significance of ITGAV variants. (A–C) Pedigrees of (A) Family 1, (B) Family 2, and (C) Family 3. Symbols: unknown genotype, ‘‘E?’’; fetus, triangle. (D) Cartoon of Integrin αV protein domain structure demonstrating protein-coding and splicing defects. TM, transmembrane. (E) Sashimi plots of RNA sequencing from fibroblasts demonstrating the novel splice acceptor site in ITGAV exon 4 of P1 (red tracks) compared with healthy control (HC, blue tracks). y axis: number of reads in log2 scale; x axis: genomic location. Only the canonical transcript model and junctions with ≥5 reads are shown. (F and G) Immunoblotting of Integrin αV protein level (n = 3) (F) and ITGAV mRNA by qPCR in fibroblasts derived from an HC or P1 (G). GAPDH was used as a loading control. qPCR was normalized to GAPDH, normalized to HC fibroblasts. (n = 3, paired t test, P < 0.01, error bars, mean ± SEM). (H and I) Immunoblotting (n = 3) (H) and Flow cytometry (I) of Integrin αV and Integrin β3 surface expression in lymphoblastoid cell lines derived from HC and P2 (n = 3). (J) Ca2+ ion (depicted as a white circle) coordination by the Asp379 Integrin αV sidechain mutated in P2, as well as the sidechains of Asp381, Asp383, and Asp387, are shown (black dashes). Stabilizing hydrogen bonds between backbone atoms and the Integrin αV Asp379 sidechain are also shown (magenta dashes). Models generated from PDB: 4G1M. (K) Spinning disc confocal microscopy of Integrin αV-FLAG alone (left) or co-expressed with Integrin β3 (right) in HeLa cells (n = 3). (L) Sashimi plots of RNA-sequencing data showing alternative ITGAV splicing of exon 16 (left) and exon 23 (right) in the mother (blue tracks), F3 (red tracks), and unrelated control (light yellow tracks). Left panel also indicates skipping and a novel cryptic acceptor site in exon17 in ITGAV (arrow). y axis: number of reads. x axis: genomic location. Only the canonical transcript model and junctions with ≥5 reads are shown. Source data are available for this figure: SourceData F2.