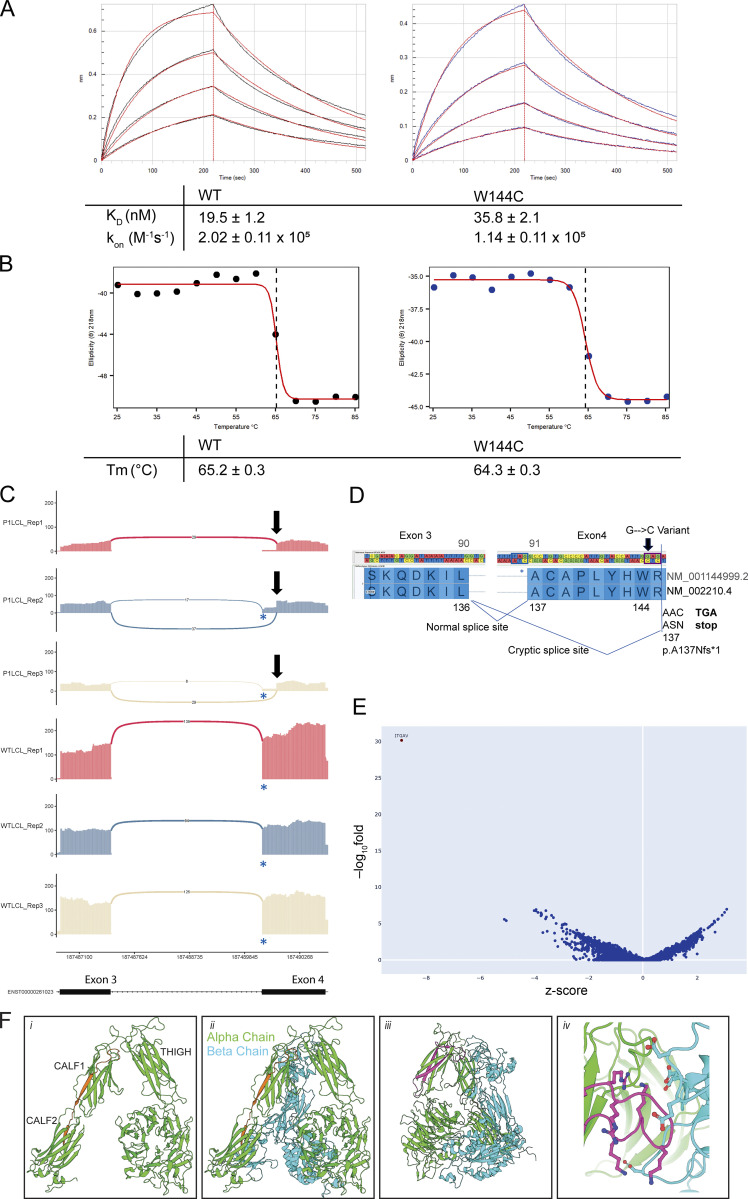
Figure S1.
Biophysical properties of recombinant ITGAV and additional RNA sequencing. (A) Representative BLI curves of WT and W144C proteins binding to biotinylated TGF-β3 peptide. KD, kon, and koff, are the equilibrium dissociation constant, on rate, and off rate, respectively. Values are indicated along with standard error; n = 3. (B) CD melting curves were observed at 218 nm for WT and W144C proteins. Dotted lines at the inflection point indicate melting temperature (Tm). (C) Sashimi plots showing the novel splicing site in ITGAV exon 4 (only junctions with ≥5 reads are shown), generated by ggsashimi. A blue * indicates the canonical splice acceptor site while a black arrow marks the novel splice site in exon 4. Three replicates are shown using a lymphoblast cell lines derived from P1 (top) and an unrelated healthy control (WT, bottom). y axis: number of reads. x axis: genomic location. Only the canonical transcript model is shown. (D) Cartoon demonstrating the abnormal splicing junction that occurs between exons 3 and 4 of ITGAV, visualized using GenomeBrowse, because of the c.432G > C variant identified in P1. ITGAV amino acid numbering is shown for the indicated two transcript IDs. The canonical splice acceptor site is highlighted using a blue box while the novel splice acceptor site is shown using a black box. The position of the variant in P1 is marked by an arrow. The new reading frame of the novel splice site gives rise to a premature stop codon. (E) Volcano plot showing differentially expressed genes from RNA-sequencing data comparing fetus (F3) derived fibroblasts versus 100 controls. x axis: z-score. y axis: −log10fold. (F) Structural modeling of inframe deletions identified in Family 3. The 27 amino acid in-frame deletion of residues S750 to G776 spanning the CALF1 and CALF2 domains (indicated in orange) would (i) invert the CALF1 domain resulting in (ii) the misalignment of the alpha and beta chains. The 44 amino acid in-frame deletion of residues F503 to S546 within the THIGH domain (iii) would ablate an (iv) electronegative loop interacting with an electropositive pocket on the alpha chain. Models generated from PDB: 4G1M.