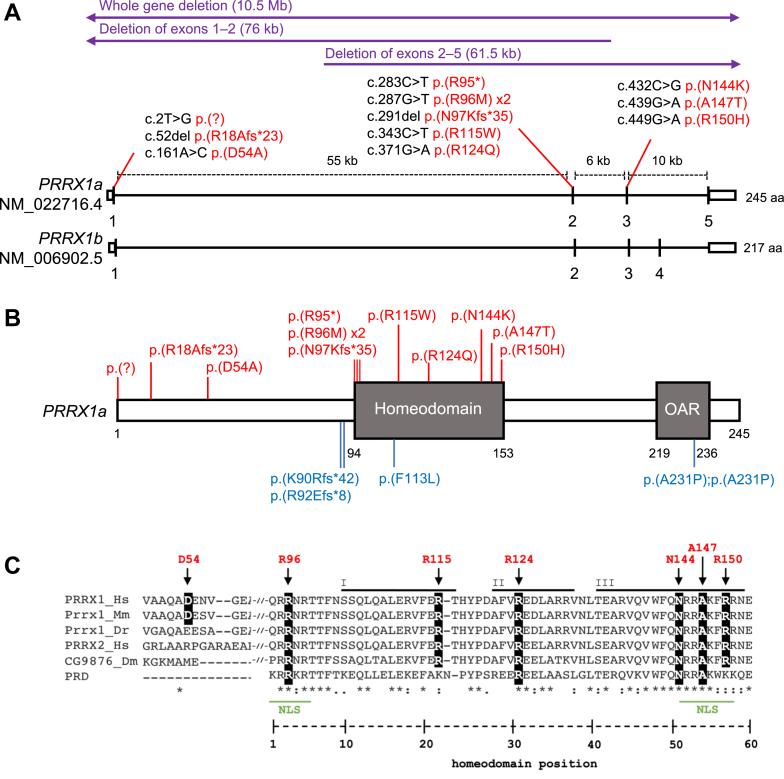
Figure 1.
Structure, conservation, and variants in PRRX1. A. A schematic of the exon structure (exons 1-5) of human PRRX1a (245 aa) and PRRX1b (217 aa). Alternative splicing of the final exon results in an OAR (otp, aristaless, rax) domain present in PRRX1a, which is absent in PRRX1b. Variants identified in patients with craniosynostosis are highlighted in red with the purple lines above indicating the 3 deletions identified in independent patients. Arrows indicate that the deletion extends beyond the PRRX1 gene. B. A schematic representation of the PRRX1a protein (P54821) showing the position of the homeodomain and the OAR domain. Variants identified in this study are highlighted in red, whereas variants reported in patients with agnathia-otocephaly are shown in blue. The only homozygous variant reported is p.(A231P) identified in a patient with agnathia-otocephaly. C. Conservation of the amino acids surrounding D54 (left) and the 60 amino acids of the homeodomain (right). Sequences correspond to human PRRX1a and PRRX2 (PRRX1_Hs and PRRX2_Hs, respectively), mouse (Prrx1_Mm), zebrafish (Prrx1_Dr), and Drosophila (CG9876_Dm), and the consensus sequence for the PRD class of homeodomain-containing proteins. The arrows indicate the position of the missense substitutions identified in this study. The 3 alpha helices of the homeodomain are also highlighted (I, II, and III). Two predicted nuclear localization sequences (NLS) are annotated in green. Asterisks (∗) represent complete conservation across all alignments, a colon (:) represents aligned residues with similar biochemical properties, and the period (.) denotes conservation between groups with weakly similar properties. aa, amino acid.