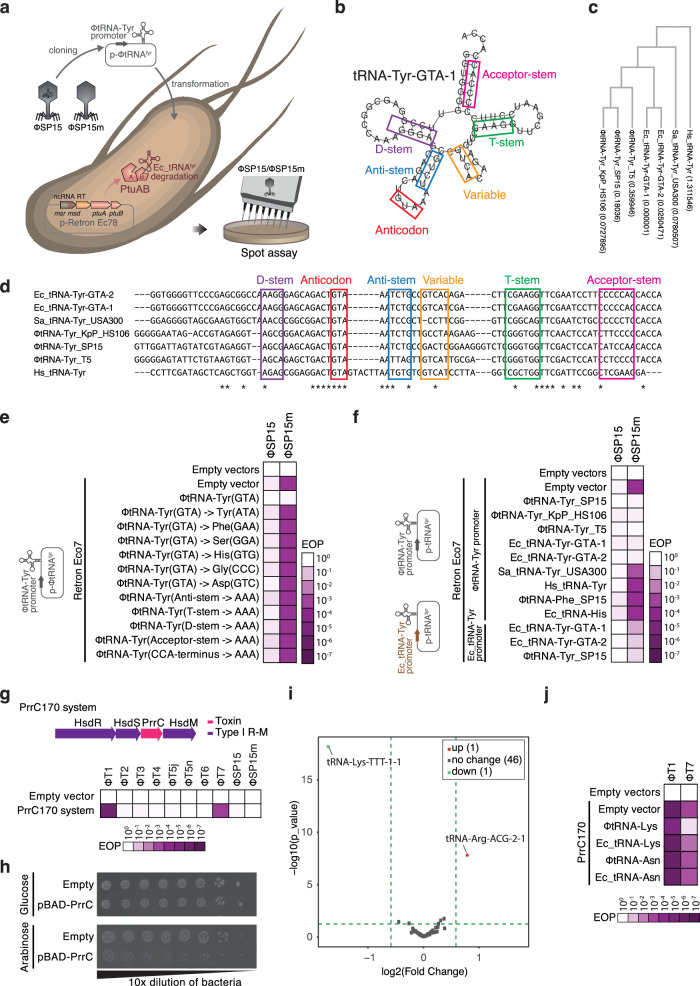
Fig. 3. Supplementation of phage-derived tRNA is strategy employed by phage to evade defense systems.
a Simplified depiction of the method used to evaluate tRNA complementation on bacteria carrying retron-Eco7. The complementation of tRNA was performed in trans by expressing the tRNA under either the phage tRNATyr promoter or the bacterial tRNATyr promoter and introducing it into bacteria carrying retron-Eco7. b RNAFold42-based structural prediction of tRNATyr from E. coli (Ec-tRNA-GTA-1). The predicted secondary structure of the tRNA is highlighted with colored box lines: D-stem (purple), anticodon loop (red), anticodon stem (blue), variable loop (orange), T-stem (green), and acceptor stem (pink). c Phylogenetic tree of the tRNATyr used for the complementation experiment. The DNA alignment was performed using ClustalW43, and the tree was generated using the bootstrap maximum likelihood method. The value inside the brackets indicates the bootstrap score. d Sequence alignment of tRNATyr from T5 (ΦtRNA-Tyr_T5), SP15 (ΦtRNA-Tyr_SP15), Klebsiella phage KpP_HS106 (ΦtRNA-Tyr_KpP_HS106), S. aureus (Sa_tRNA-Tyr_USA300), Homo sapiens (Hs_tRNA-Tyr), and E. coli tRNATyr (Ec-tRNA_Tyr-GTA-1 or Ec-tRNA_Tyr-GTA-2). Based on the predicted secondary structure of tRNATyr from E. coli, the loop, stem, and anticodon sequences are all highlighted using colored boxes. e Heatmap of phage EOP illustrating the mutation in different stem loops and changing the anticodon sequence of ΦtRNA-Tyr_SP15, which abolished the tRNA ability to rescue the phage from retron-Eco7. SP15 and SP15m were used in the phage assay. f Heatmap of phage EOP illustrating the expression of tRNATyr from different phages (ΦtRNA-Tyr_T5 and ΦtRNA-Tyr_KpP_HS106) or from E. coli in rescuing phages from retron-Eco7 in a promoter-dependent manner. The tRNA was expressed under either the phage tRNATyr promoter (ΦtRNA-Tyr promoter) or the E. coli tRNATyr promoter (Ec_tRNA-Tyr promoter). Hs_tRNA-Tyr, Sa_tRNA-Tyr_USA300, and E. coli tRNAHis (Ec_tRNA-His) were unable to rescue phages from retron-Eco7. The photograph of the spot assay and the phage count graph of the heatmaps in (e) and (f) are presented in Supplementary Fig. 10a–d. Source data are provided as a Source Data File. g Heatmap of phage EOP illustrating the antiphage function of the PrrC170 anticodon nuclease (named after isolate number 170 of carbapenem-resistant Klebsiella quasipneumoniae) against at least two phages, T1 and T7. The PrrC170 system comprises PrrC and an associated restriction-modification system type I, cloned in pLG001 plasmid3 under its native promoter. The photograph of the spot assay and the phage count graph are available in Supplementary Fig. 13a, b. Source data are provided as a Source Data File. h Growth arrest observed in bacteria expressing the PrrC toxin. The prrC gene was cloned under the pBAD inducible plasmid. i Volcano plot depicting tRNA sequencing of bacteria carrying the pBAD-PrrC toxin. The fold change was calculated based on the total tRNA expression level in bacteria under induction (arabinose added) versus repression (glucose added). The log2(Fold Change) represents the difference in means between two groups, calculated as PrrC-induce_CPM minus PrrC-repress_CPM. The statistical significance was determined using the p-value from the exact test based on a negative binomial distribution. No adjustments for multiple comparisons were made. j Heatmap of phage EOP illustrating the complementation of tRNALys from the SP15 (ΦtRNA-Lys) phage in rescuing phages from the PrrC170 defense system. Complementation of E. coli tRNALys (Ec_tRNA-Lys), E. coli tRNAAsn (Ec_tRNA-Asn), tRNAAsn from SP15 (ΦtRNA-Asn) did not rescue phage from PrrC170. The complementation was performed by expressing the tRNA in trans under phage tRNA promoter and introducing it into bacteria carrying PrrC170. The phage count graph is provided in Supplementary Fig. 13c. Source data are provided as a Source Data File.